Difference between revisions of "Journal:Characterization of trichome phenotypes to assess maturation and flower development in Cannabis sativa L. by automatic trichome gland analysis"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 19: | Line 19: | ||
|website = [https://www.sciencedirect.com/science/article/pii/S2772375522000764 https://www.sciencedirect.com/science/article/pii/S2772375522000764] | |website = [https://www.sciencedirect.com/science/article/pii/S2772375522000764 https://www.sciencedirect.com/science/article/pii/S2772375522000764] | ||
|download = [https://www.sciencedirect.com/science/article/pii/S2772375522000764/pdfft?md5=bcc814b8233cdb0430097fbeca72524d&pid=1-s2.0-S2772375522000764-main.pdf https://www.sciencedirect.com/science/article/pii/S2772375522000764/pdfft] (PDF) | |download = [https://www.sciencedirect.com/science/article/pii/S2772375522000764/pdfft?md5=bcc814b8233cdb0430097fbeca72524d&pid=1-s2.0-S2772375522000764-main.pdf https://www.sciencedirect.com/science/article/pii/S2772375522000764/pdfft] (PDF) | ||
}} | |||
{{ombox | |||
| type = content | |||
| style = | |||
| text = This article contains rendered mathematical formulae. You ''may'' require the [https://chrome.google.com/webstore/detail/tex-all-the-things/cbimabofgmfdkicghcadidpemeenbffn?hl=en TeX All the Things] plugin for Chrome or the [https://addons.mozilla.org/en-US/firefox/addon/native-mathml/ Native MathML] add-on and [https://developer.mozilla.org/en-US/docs/Web/MathML/Fonts fonts] for Firefox if they don't render properly for you. | |||
}} | }} | ||
{{ombox | {{ombox | ||
| Line 121: | Line 126: | ||
DO-U-Net achieves instance segmentation by subtracting the predicted edges of trichome glands from the trichome gland mask, such that individual glands are isolated. We apply the same loss function and training parameters as outlined in the DO-U-Net method, only making minor adjustments to edge and mask post processing such that adjacent trichome glands are separated by single-pixel-width lines. We noted that the original implementation of DO-U-Net tends to shrink the size of detected trichome glands due to the edge subtraction operation, thus we incorporated a custom post processing method using Voronoi cells to mitigate this effect (Fig. A2, Appendix). The DO-U-Net training and testing data was a manually annotated subset of the data described earlier, consisting of 156 (136:20 training:testing) images drawn uniformly with respect to strain and observation time to construct a balanced segmentation dataset (Fig. A1, Appendix). We implemented DO-U-Net in PyTorch, using the built in ADAM optimizer. To ensure the network generalized well to unseen data, we augmented the inputs at training time using standard augmentation procedures: random horizontal or vertical flipping, random rotation, and random color jitter. Training time was approximately two hours and was terminated after 100 epochs running on an Nvidia 1080ti GPU with 12 GB memory. The network was trained on 512×512 pixel image patches randomly cropped from input images. At inference, output mask patches and edge patches were stitched back together to compute mask and edge predictions for whole bract tissue images, followed by application of our post processing procedure. We refer the reader to the Appendix where supplementary figures describe the post-processing steps in detail. | DO-U-Net achieves instance segmentation by subtracting the predicted edges of trichome glands from the trichome gland mask, such that individual glands are isolated. We apply the same loss function and training parameters as outlined in the DO-U-Net method, only making minor adjustments to edge and mask post processing such that adjacent trichome glands are separated by single-pixel-width lines. We noted that the original implementation of DO-U-Net tends to shrink the size of detected trichome glands due to the edge subtraction operation, thus we incorporated a custom post processing method using Voronoi cells to mitigate this effect (Fig. A2, Appendix). The DO-U-Net training and testing data was a manually annotated subset of the data described earlier, consisting of 156 (136:20 training:testing) images drawn uniformly with respect to strain and observation time to construct a balanced segmentation dataset (Fig. A1, Appendix). We implemented DO-U-Net in PyTorch, using the built in ADAM optimizer. To ensure the network generalized well to unseen data, we augmented the inputs at training time using standard augmentation procedures: random horizontal or vertical flipping, random rotation, and random color jitter. Training time was approximately two hours and was terminated after 100 epochs running on an Nvidia 1080ti GPU with 12 GB memory. The network was trained on 512×512 pixel image patches randomly cropped from input images. At inference, output mask patches and edge patches were stitched back together to compute mask and edge predictions for whole bract tissue images, followed by application of our post processing procedure. We refer the reader to the Appendix where supplementary figures describe the post-processing steps in detail. | ||
===Trichome phenotype classification=== | |||
A k-NN classifier was trained on manually labeled trichome glands (Fig. 5). For each strain, 10 glands from the classes representing "clear," "milky," and "brown" were delineated manually. Each segmented gland was transformed from the sRGB to the CIELAB color space to disentangle color from luminosity, reducing the effect of shadows on classifier accuracy. Features were extracted by taking the mean of the L, a, and b color channels, and the mean of a Hessian (H) filter applied to the chroma: <math>c = \sqrt{a^{2} + b^{2}}</math> | |||
[[File:Fig5 Sutton SmartAgTech2023 3.jpg|1200px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1200px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 5.''' Color figure of k-NN classifier. Left: Ten clear, milky, and brown trichomes (gland columns l-3 respectively) used to train the k-NN classifier for strain GDB, and the resulting t-SNE clustering of training features. Right: At inference, the k-NN classifier Ⓐ extracts features and Ⓑ predicts the probability of each gland instance belonging to class clear, milky, brown. A k-NN classifier is constructed for each strain, here we only show the classifier for GDB for brevity.</blockquote> | |||
|- | |||
|} | |||
|} | |||
The Hessian filter is responsive to fine lines and details observed in clear trichomes but obscured in milky trichomes. For inference, k was set to 5, and the votes of training points were weighted by inverse distance. The mean and standard deviation calculated from the training data were applied to test data to standardize features at inference. | |||
===Analysis of trichome morphology=== | |||
Changes in trichome morphology were assessed by computing metrics describing connected components in the post-processed semantic segmentation maps predicted by the DO-U-Net. Connected components are groups of contiguous foreground pixels representing a trichome head gland. The average distance to the nearest trichome gland (ADT), trichome gland density (trichome glands per mm<sup>2</sup>), trichome diameter (in microns), trichome clumping fraction (CF), and roundness were computed from the data for each strain and phenotype where phenotype specific metrics were meaningful. The ADT was calculated as the mean of the distance transform describing the average distance to the nearest trichome gland over the area of the bract tissue. Trichome density was computed by counting the number of trichome instances and dividing by the pixel area of the bract tissue, then scaling by the spatial pixel size of 2 µm. Trichome diameter was computed as the diameter of a disk with the equivalent pixel area to each trichome gland instance. CF was calculated by comparing the number of trichome gland instances detected in each bract image when connectivity was defined as four-pixel or eight-pixel. In other words, trichome gland instances that were delineated by a single pixel line after post processing were deemed separate by four-connectivity but connected by eight-connectivity. A comparison of the two trichome gland counts indicated how many trichomes were clumped together in the image. | |||
The CF is expressed as: <math>\frac{\left( {N_{4} - N_{8}} \right)}{N_{4}}</math>, for each image where N is the number of trichome instances detected by the connectivity denoted by the subscript (Fig. A3, Appendix). | |||
Roundness was calculated by the circularity metric: <math>4\pi \cdot \frac{A_{gland}}{Perimeter_{gland}^{2}}</math> | |||
===Statistical analysis=== | |||
Data were subjected to independent two-sided T-tests for significance in validation experiments (Milstop and disease induced stunting) at ''p'' < 0.05. For trichome head diameter results and the morphological metrics ADT, density, CF, and roundness, the Seaborn plotting library visualized means, standard deviations of the mean, and first or second order regression lines. For regression lines, the 95% confidence intervals of trend lines were visualized to assess whether the confidence intervals of the trend lines overlapped (Figs. 8 and 15, later in this article). Confidence intervals that did not overlap were interpreted as being trendlines of significantly different groups. | |||
===Validation=== | |||
We validated our algorithm using a hybrid approach, combining manual assessment of gland segmentation and classification accuracy, and experimental assessment of algorithm predictions compared to prior biological knowledge of the cannabis flower. Pixelwise metrics, although useful for determining performance discrepancies between automatic methods, have less meaning when assessing the performance of an algorithm in the context of the biological domain being investigated. Therefore, we turn to the recommendation of Sbalzarini [20], who argued that the most pragmatic method of validating a bio-imaging algorithm is to test predictions against prior knowledge from the same domain as the research question. We conducted two validation experiments: the detection of stunted trichome maturation as predicted by trichome diameter when flowers were subjected to viroid disease stress, and the detection of trichome browning when flowers were sprayed with an alkaline chemical treatment. | |||
==Results== | |||
We manually inspected the accuracy of the predicted segmentation masks on a withheld test set of bract images. We observed a low false positive rate and satisfactory instance segmentation of whole trichome gland heads (Fig. 6). Glands that detached from stalks and released their contents to form amorphous blobs of resin were not detected. The accuracy of the k-NN classifier was verified via manual assessment of bract tissue images at different flowering time points to confirm trichome head phenotype predictions correctly described trichome gland development from transparent sessile and pre-stalked capitate (clear) to capitate stalked abundance (milky) prior to harvest time until trichome gland senescence (brown). | |||
[[File:Fig6 Sutton SmartAgTech2023 3.jpg|800px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 6.''' Color figure of segmented and classified trichomes glands. Top to bottom: immature, mature and senescent bract tissue of strain PK. Glands are circled according to their classification: clear (blue), milky (green), or brown (red). Zoomed in patches show detailed segmentation and classification results.</blockquote> | |||
|- | |||
|} | |||
|} | |||
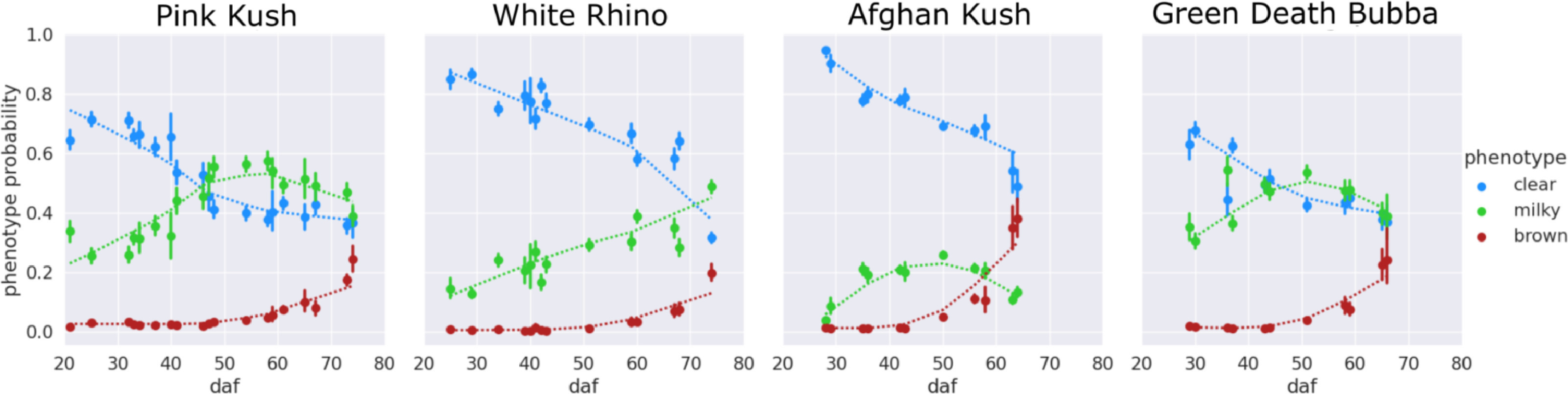
An examination of the clear-milky-brown results of all four cannabis strains over flowering time showed that in general, trichome browning was absent until approximately 50 days after flowering (daf) (Fig. 7). There was a rapid increase in the prevalence of brown trichome heads at 65–75 days of the flowering period. By comparison, the clear trichome heads were most abundant at day 25 of the flowering period, and began to steadily decline up to the end of the experiment (harvest time, day 65–75 of the flowering period, depending on the strain). The trichome heads displaying the milky phenotype began to gradually increase in prevalence in each strain, with a peak at day 55 in PK, at day 50 in strains AK and GDB, and a maximum at day 70 in WR. A significant difference in trichome gland head phenotype was not observed when assessed by dissection morphology, as outlined in Fig. 3. | |||
[[File:Fig7 Sutton SmartAgTech2023 3.jpg|1000px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1000px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 7.''' Color figure of clear, milky, brown phenotype probability. Local regression trend lines (dotted) of clear (blue), milky (green) and brown (red) phenotype probability. Points are means of the observed trichomes, with error bars showing the standard deviation of the mean. Results should be interpreted as the probability of a randomly chosen trichome, observed at a time during the flowering period, belonging to a phenotype class clear, milky, or brown. Local regression lines are for visualizing the phenotype trend over the flowering period.</blockquote> | |||
|- | |||
|} | |||
|} | |||
==References== | ==References== | ||
Revision as of 17:03, 7 June 2023
| Full article title | Characterization of trichome phenotypes to assess maturation and flower development in Cannabis sativa L. (cannabis) by automatic trichome gland analysis |
|---|---|
| Journal | Smart Agricultural Technology |
| Author(s) | Sutton, D.B.; Punja, Z.K.; Hamarneh, G. |
| Author affiliation(s) | Simon Fraser University |
| Primary contact | Email: darrens at sfu dot ca |
| Year published | 2023 |
| Volume and issue | 3 |
| Article # | 100111 |
| DOI | 10.1016/j.atech.2022.100111 |
| ISSN | 2772-3755 |
| Distribution license | Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International |
| Website | https://www.sciencedirect.com/science/article/pii/S2772375522000764 |
| Download | https://www.sciencedirect.com/science/article/pii/S2772375522000764/pdfft (PDF) |
|
|
This article contains rendered mathematical formulae. You may require the TeX All the Things plugin for Chrome or the Native MathML add-on and fonts for Firefox if they don't render properly for you. |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Cannabis (Cannabis sativa L.) is cultivated by licensed producers in Canada for medicinal and recreational uses. The recent legalization of this plant in 2018 has resulted in rapid expansion of the industry, with greenhouse production representing the most common method of cultivation. Female Cannabis plants produce inflorescences that contain bracts densely covered by glandular trichomes, which synthesize a range of commercially important cannabinoids (e.g., tetrahydrocannabinol [THC] and cannabidiol [CBD]), as well as terpenes. Cannabinoid content and quality varies over the eight-week flowering period to such an extent that the time of harvest can significantly impact product quality. Cannabis flower maturation is accompanied by a transition in the color of trichome heads that progresses from clear to milky to brown (amber) and can be seen visually using low magnification. However, the importance of this transition as it impacts quality and describes maturity has never been investigated.
To establish a relationship between trichome maturation and trichome head color changes (phenotype), we developed a novel automatic trichome gland analysis pipeline using deep learning. We first collected a macro-photography dataset based on four commercially grown Cannabis strains, namely Afghan Kush, Green Death Bubba, Pink Kush, and White Rhino. Images were obtained in two modalities: conventional macroscopic light photography and macroscopic UV induced fluorescence. We then implemented a pipeline where the clear-milky-brown heuristic was injected into the algorithm to quantify trichome phenotype progression during the eight-week flowering period. A series of clear, milky, and brown phenotype curves were recorded for each strain over the flowering period that were validated as indicators of trichome maturation and corresponded to previously described parameters of trichome development, such as trichome gland head diameter and stalk elongation. We also derived morphological metrics describing trichome gland geometry from deep learning segmentation predictions that profiled trichome maturation over the flowering period.
We observed that mature and senescing trichomes displayed fluorescent properties that were reflected in the clear, milky, and brown phenotypes. Our method was validated by two experiments where factors affecting trichome quality and flower development were imposed, and the effects were then quantified using the deep learning pipeline. Our results indicate the feasibility of automated trichome analysis as a method to evaluate the maturation of female flowers cultivated in a highly variable environment, regardless of strain. These findings have broad applicability in a growing industry in which cannabis flower quality is receiving increased circumspection for medicinal and recreational uses.
Keywords: Cannabis, trichomes, deep learning, phenotype, fluorescence, precision agriculture
Introduction
Cannabis (Cannabis sativa L.) is valued for its medicinal use in every continent except Antarctica, and many countries have established the legal framework for the [Cannabis cultivation|cultivation]] and sale of recreational-cannabis-derived products. [1] Drug-producing Cannabis strains are characterized by large female inflorescences (flowers) that bear a cluster of pistils surrounded by bracts, which produce large numbers of glandular trichomes where cannabinoids and terpenes are synthesized. [2] Cannabis plants require approximately eight weeks to mature prior to harvest, after which inflorescences are dried for sale in their natural form or processed for value-added products (i.e., edibles, cosmetics, extracted oils). In order to ensure optimal Cannabis quality, it is imperative to identify the stage at which the inflorescences are at the point of prime maturation and hence potency. There are currently no scientifically based methods to predict maturation of inflorescences, and consequently harvests are performed on a calendar basis, i.e., when plants have attained seven to eight weeks of growth. Enacting methods to better assess trichome maturation can lead to improvements in quality assurance for the Cannabis industry.
In this work, we study Cannabis trichome gland head phenotypes as flowers mature and visual trichome changes occur, such as a progression in color of the heads of trichomes from clear to milky to brown. Although these phenotypes have been suggested as a visual heuristic for harvest timing, little scientific work describes them during flower development and trichome maturation. Previous work supports the idea that browning of trichome heads is associated with quality degradation in dried Cannabis [3], but the shelf life of dried Cannabis [4] and progressive trichome browning in storage makes extrapolation of results to fresh Cannabis tissue unpredictable. In order to understand the role of trichome phenotypes during trichome maturation, it is necessary to obtain measurements in situ during flower development.
We describe an automatic computational method that was built to extract trichome phenotype and morphology metrics during Cannabis flower development from macroscopic photographs. To reduce uncertainty related to the chronology of observations, our method was implemented in a commercial greenhouse such that the time delay between excision and photography was minimized. By implementing recent advances in computer vision, we show our automatic method can be used to define trichome maturation in multiple Cannabis strains in high-throughput applications without repeated fine tuning.
Related work
Morphology of cannabis trichomes
Trichomes are ubiquitous structures in the plant kingdom. Many essential oil-bearing plants possess glandular trichomes that have important commercial value. [5] Trichomes can also consist of plant hairs lacking a gland, with a spike-like appearance. [6] In hemp and drug-producing Cannabis plants, Hammond and Mahlberg [7] studied trichome types using scanning electron microscopy (SEM) and described three types: bulbous, sessile, and capitate stalked. They subsequently described the morphological development of capitate stalked trichomes, observing stalk elongation and resin accumulation in the gland on top of the secretory cells. [8] These contributions laid the groundwork for the understanding of cannabinoid synthesis and storage in planta. Mahlberg and Kim [9] further characterized the formation of the trichome gland cuticle and proposed a working model of cannabinoid biosynthesis, hypothesizing that tetrahydrocannabinolic acid (THCA) is formed outside of plant cells in the storage cavity of the trichome gland. [10] Sirikantaramas et al. [11] confirmed this hypothesis by localizing the enzymes necessary for cannabinoid synthesis to the storage cavity, determining that cannabinoids are formed from a ring of secretory cells. Livingston et al. [12] further characterized the development of cannabis trichomes and observed that capitate stalked trichomes develop from sessile type trichomes during the flowering period. The development of trichomes from the sessile to cannabinoid-rich capitate stalked type was accompanied by a shift in autofluorescence of the gland contents from green to blue.
Trichome maturation
The color transition of trichome heads from clear to milky to brown is often used to manually approximate the stage of maturation, with milky representing the optimal state and brown indicating over-maturation. [10,13] However, the relationship between trichome head color and cannabis flower development has not been previously investigated. Previous work has shown a possible negative correlation between trichome head browning and cannabinoid content. Turner et al. [14] were the first to show reduced cannabinoid content in senescent (brown) capitate stalked glands, although statistical analysis was not reported. Cannabis samples acquired from law enforcement and manually assessed for trichome browning on a linear scale had a lower THCA and increased cannabinolic acid (CBNA) content compared to Cannabis samples with clear or milky trichomes. [3] Reduced tetrahydrocannabinol (THC) content in senescent trichomes glands was also reported by Mahlberg and Kim. [10] There is currently a lack of prior reports describing trichome gland phenotype in relationship to Cannabis flower development in situ in Cannabis.
Automatic assessment of plant trichomes
The microscopic nature of trichomes complicates manual observation, but recent advances in automatic methods to assess trichomes have enabled increasingly powerful analyses. Glandular trichomes of tomato were sorted by autofluorescent flow cytometry to separate young and mature trichomes prior to transcriptome analysis of the gland contents. [15] Failmezger et al. [16] estimated the 3D leaf surface of Arabidopsis thaliana from 2D microscopy image stacks to characterize trichome growth patterns. Mirnezami et al. [17] investigated image processing to automatically count soybean trichomes from microscopy images. Deep learning has also been applied to trichome analysis to estimate the "hairiness" of cotton leaves as a metric of harvest readiness. [18] No previous studies have assessed trichome maturation in Cannabis using these or similar approaches; thus, metrics that could be used to predict when Cannabis inflorescences have achieved a maturation stage indicative of optimal cannabinoid content have not been established.
Contributions
In this work, we document changes in trichome gland head phenotypes over the flowering period and compare observations of the trichome glands and the supporting bract tissue. We present a computer vision pipeline to automatically segment and classify trichome gland head phenotypes and validate the pipeline by experiments that induce trichome degradation and alter trichome development. We also explore novel fluorescent trichome gland head traits induced under UV irradiation that were observed from whole glands, as opposed to previous studies that explore fluorescence in a confocal manner. [12,31] We acquired data using conventional optics such that our analysis was carried out in situ and yielded same-day results. We discuss our results in the context of trichome development and Cannabis flower maturation in four Cannabis strains (genotypes) and conclude with comments on the feasibility of on-site imaging technology to automate quality aspects during the Cannabis cultivation process in the greenhouse. In the following sections, we first describe plant materials, plant propagation, and computational analysis, then present results and conclude with the interpretation of results in the discussion.
Materials and methods
Plant materials
This research was conducted in a Health-Canada-approved facility in which the plants were grown in a commercial greenhouse using a hydroponic method of cultivation. There were four strains (genotypes) studied that were grown during the winter months (October-April 2021; strains Pink Kush or "PK," and White Rhino or "WR") or during the summer months (May-September 2020; strains Afghan Kush or "AK," and Green Death Bubba or "GDB"). During the winter period, plants were provided with supplementary lighting to achieve 1,600 uEin/m2 of photosynthetically active radiation (PAR) using sodium lamps emitting a wavelength centered at 590 nm. During the summer period, supplementary lighting was provided where needed on overcast days to achieve the required PAR.
All plants were initiated from cuttings taken from stock (mother) plants of the four strains. The cuttings were dipped in rooting powder (containing indoleacetic acid) and inserted into 2.5 cm rockwool cubes, which were placed in trays in a propagation room under high relative humidity (80%) and a temperature range of 23–27 °C. After two weeks, when cuttings had rooted, they were placed into wells cut into 10 cm3 rockwool blocks and placed on a greenhouse bench for an additional two weeks to acclimatize and resume vegetative growth. Following this, they were transferred into large cocofibre blocks, one plant per block, and placed in a large flowering room where the photoperiod was adjusted to 12-hour lighting and 12-hour darkness to induce flowering. The water and nutrient regimes were adjusted according to commercial growing requirements to ensure adequate growth. The bags were arranged in rows, with two rows parallel to each other, and spacing of 0.5 meters between plants. The plants were trained and supported by wire mesh netting that allowed the developing inflorescences to grow upright. Pruning of leaves and training of plants were conducted according to commercial growing requirements.
For experimental observations where inflorescences were photographed, each row of plants in the flowering room was divided into 10 zones, each containing up to 10 plants for observation such that no inflorescence was excised from the same plant twice. The zones were located in the center of the greenhouse to ensure uniform growing conditions. Plants were not treated with any foliar sprays that could influence trichome development. However, for one group of plants, a spray of potassium bicarbonate (Milstop) was applied to Cannabis flowers of strain PK. Two plants each were sprayed with 100 ml of 2.5 g/L Milstop at five weeks and six weeks after flowering using a hand-held sprayer. Two control plants were misted with water. Images were collected the following week (flowering week seven) from these plants. For segmentation validation, plants from strain PK, which were visually stunted due to infection by a viroid disease, were compared to healthy plants. Images were collected from a stunted plant and two healthy control plants at flowering week seven.
Data collection
Images of inflorescences were obtained from plants that were in the third to ninth week of flowering. Images were collected weekly from the onset of visible trichomes (week three) to flower senescence (weeks eight or nine, depending on strain). The inflorescences selected for photography were situated on the terminal apical branches of plants. Individual buds were carefully removed using pruning shears and transported to a designated room for image collection. The individual buds were separated into segments representing apical, mid-, and bottom regions, and bract tissue samples were selected from both sides (Fig. 1). To capture the images of trichomes, both visible lighting (range of 400–700 nm provide by fluorescent lamps) and ultraviolet (centered at 350 nm) were used. The latter was achieved by using Baader UV-pass filters, as described in the next subsection. Each image was labelled according to location of the flower sample, day of the flowering period, Cannabis strain, and illumination source. The dataset consisted of 840, 840, 720, and 720 images of strains PK, WR, AK, and GDB, respectively. The former two strains were observed for an additional week to capture trichome browning. Image resolution was 6240×4160.
|
Image collection by photography
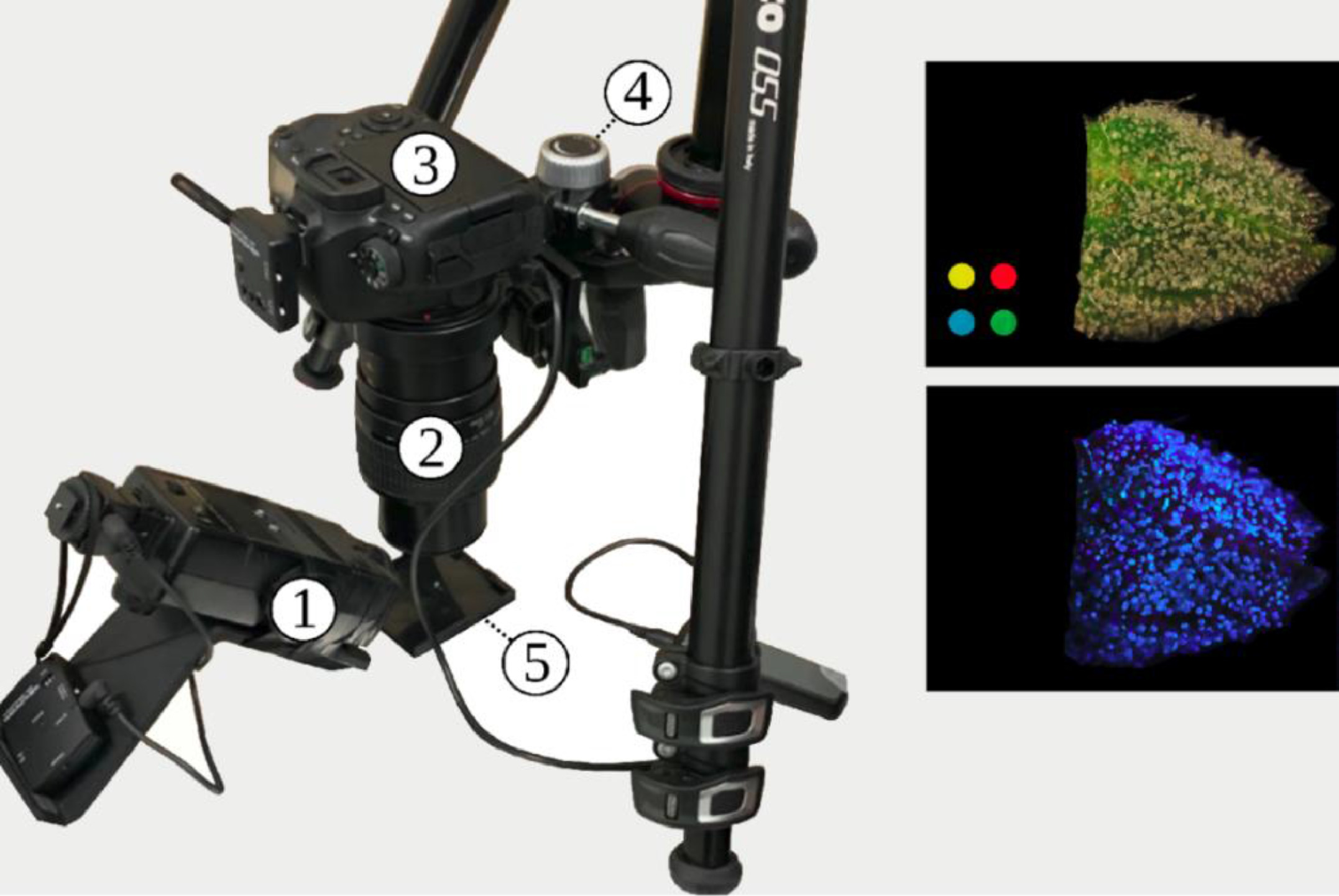
All photographs were captured with a Canon 6D Mk. II DSLR Camera (Canon, JP) fitted with a Canon MP-E 65 mm 1–5x macroscopic lens (Fig. 2). Camera configuration was as follows: Lens magnification 3x macro, f-number 16:14 conventional:fluorescent, color temperature 5500 K, sRGB color space, and exposure time of one second for both modalities. The camera was mounted on a table with vibration damping grommets. For UV excitation to photograph autofluorescence, a modified Vivitar 283 xenon flash (Vivitar, Santa Monica USA) had the stock Fresnel lens replaced with a Baader UV-pass filter (Baader-Planetarium, Munich DE) resulting in a flash centered at 350 nm and with a 60 nm bandwidth (320 – 380 nm). The single-photon fluorescence excitation wavelength of ∼350 nm used in this study is approximately equivalent to the two-photo excitation wavelength of 720 nm used by Livingston et al. [12] to characterize the fluorescence of sessile and capitate stalked trichomes of hemp. Our approach uses simpler optics such that the camera prototype can be dismantled and redeployed rapidly in situ, and the chosen fluorescence excitation wavelength is outside of the spectral sensitivity of most conventional image sensors. The Vivitar flash was synchronized to the Canon DSLR camera via Elinchrom wireless flash trigger. All images were captured with a color calibration swatch in view, and fluorescent images were captured under conditions of complete darkness.
|
Machine learning methods
Trichome gland segmentation
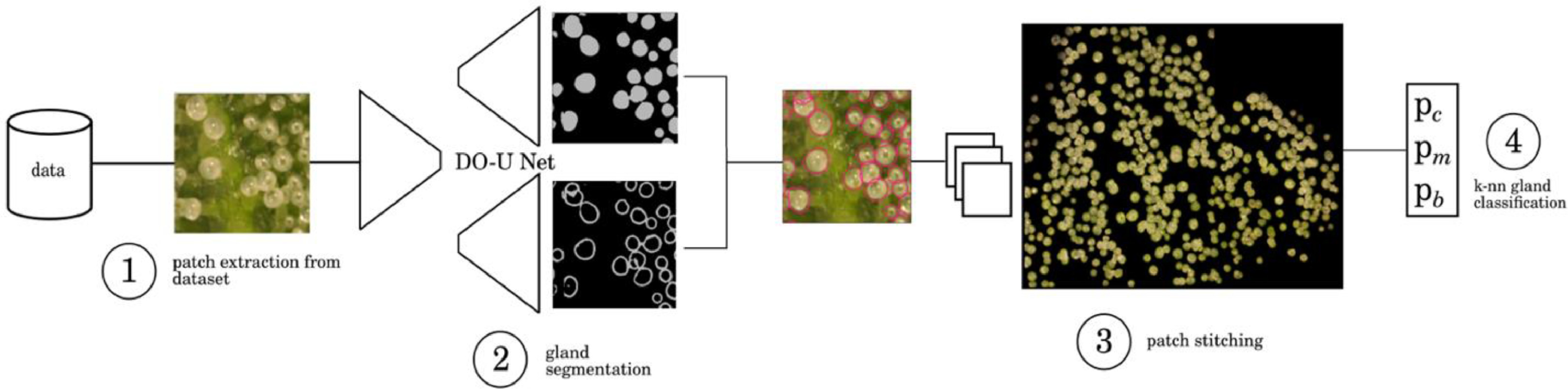
Observations were made of trichome glands (Fig. 3) that formed on the surfaces of bract tissues. Trichome glands were often partially occluded or stuck to other trichomes, complicating the segmentation of individual gland instances. Instance segmentation is a difficult problem in computer vision because it requires the algorithm to separate the foreground from the background in the image and individual components (trichome glands) of the foreground scene. We used the deep learning neural network DO-U-Net [19] designed for instance segmentation of repeated convex shapes in 2D microscopy and aerial images, where a dual decoder U-Net architecture predicts a semantic mask describing foreground membership (trichome glands), and the edges of each trichome gland in the image (Fig. 4).
|
|
DO-U-Net achieves instance segmentation by subtracting the predicted edges of trichome glands from the trichome gland mask, such that individual glands are isolated. We apply the same loss function and training parameters as outlined in the DO-U-Net method, only making minor adjustments to edge and mask post processing such that adjacent trichome glands are separated by single-pixel-width lines. We noted that the original implementation of DO-U-Net tends to shrink the size of detected trichome glands due to the edge subtraction operation, thus we incorporated a custom post processing method using Voronoi cells to mitigate this effect (Fig. A2, Appendix). The DO-U-Net training and testing data was a manually annotated subset of the data described earlier, consisting of 156 (136:20 training:testing) images drawn uniformly with respect to strain and observation time to construct a balanced segmentation dataset (Fig. A1, Appendix). We implemented DO-U-Net in PyTorch, using the built in ADAM optimizer. To ensure the network generalized well to unseen data, we augmented the inputs at training time using standard augmentation procedures: random horizontal or vertical flipping, random rotation, and random color jitter. Training time was approximately two hours and was terminated after 100 epochs running on an Nvidia 1080ti GPU with 12 GB memory. The network was trained on 512×512 pixel image patches randomly cropped from input images. At inference, output mask patches and edge patches were stitched back together to compute mask and edge predictions for whole bract tissue images, followed by application of our post processing procedure. We refer the reader to the Appendix where supplementary figures describe the post-processing steps in detail.
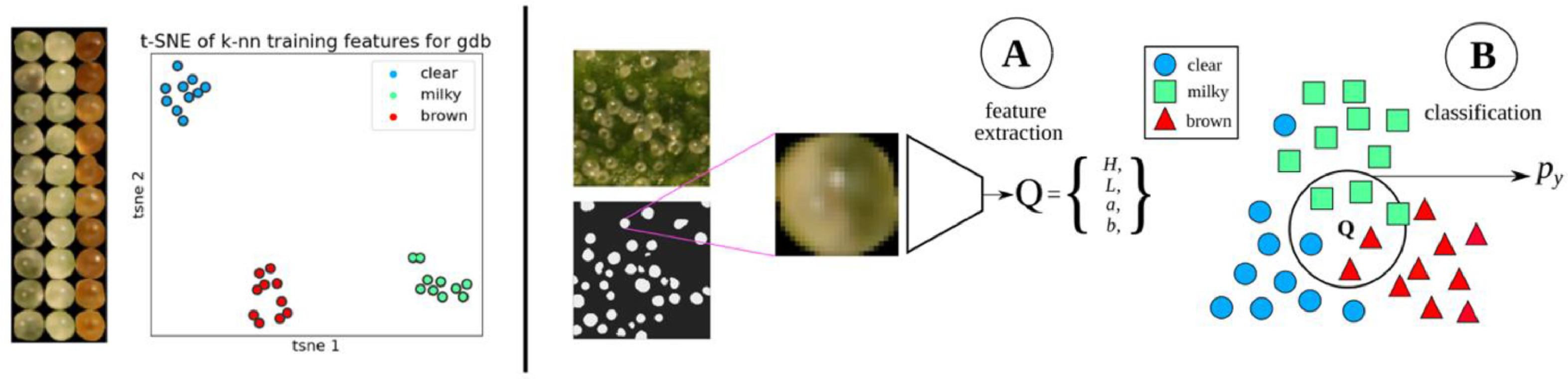
Trichome phenotype classification
A k-NN classifier was trained on manually labeled trichome glands (Fig. 5). For each strain, 10 glands from the classes representing "clear," "milky," and "brown" were delineated manually. Each segmented gland was transformed from the sRGB to the CIELAB color space to disentangle color from luminosity, reducing the effect of shadows on classifier accuracy. Features were extracted by taking the mean of the L, a, and b color channels, and the mean of a Hessian (H) filter applied to the chroma:
|
The Hessian filter is responsive to fine lines and details observed in clear trichomes but obscured in milky trichomes. For inference, k was set to 5, and the votes of training points were weighted by inverse distance. The mean and standard deviation calculated from the training data were applied to test data to standardize features at inference.
Analysis of trichome morphology
Changes in trichome morphology were assessed by computing metrics describing connected components in the post-processed semantic segmentation maps predicted by the DO-U-Net. Connected components are groups of contiguous foreground pixels representing a trichome head gland. The average distance to the nearest trichome gland (ADT), trichome gland density (trichome glands per mm2), trichome diameter (in microns), trichome clumping fraction (CF), and roundness were computed from the data for each strain and phenotype where phenotype specific metrics were meaningful. The ADT was calculated as the mean of the distance transform describing the average distance to the nearest trichome gland over the area of the bract tissue. Trichome density was computed by counting the number of trichome instances and dividing by the pixel area of the bract tissue, then scaling by the spatial pixel size of 2 µm. Trichome diameter was computed as the diameter of a disk with the equivalent pixel area to each trichome gland instance. CF was calculated by comparing the number of trichome gland instances detected in each bract image when connectivity was defined as four-pixel or eight-pixel. In other words, trichome gland instances that were delineated by a single pixel line after post processing were deemed separate by four-connectivity but connected by eight-connectivity. A comparison of the two trichome gland counts indicated how many trichomes were clumped together in the image.
The CF is expressed as: , for each image where N is the number of trichome instances detected by the connectivity denoted by the subscript (Fig. A3, Appendix).
Roundness was calculated by the circularity metric:
Statistical analysis
Data were subjected to independent two-sided T-tests for significance in validation experiments (Milstop and disease induced stunting) at p < 0.05. For trichome head diameter results and the morphological metrics ADT, density, CF, and roundness, the Seaborn plotting library visualized means, standard deviations of the mean, and first or second order regression lines. For regression lines, the 95% confidence intervals of trend lines were visualized to assess whether the confidence intervals of the trend lines overlapped (Figs. 8 and 15, later in this article). Confidence intervals that did not overlap were interpreted as being trendlines of significantly different groups.
Validation
We validated our algorithm using a hybrid approach, combining manual assessment of gland segmentation and classification accuracy, and experimental assessment of algorithm predictions compared to prior biological knowledge of the cannabis flower. Pixelwise metrics, although useful for determining performance discrepancies between automatic methods, have less meaning when assessing the performance of an algorithm in the context of the biological domain being investigated. Therefore, we turn to the recommendation of Sbalzarini [20], who argued that the most pragmatic method of validating a bio-imaging algorithm is to test predictions against prior knowledge from the same domain as the research question. We conducted two validation experiments: the detection of stunted trichome maturation as predicted by trichome diameter when flowers were subjected to viroid disease stress, and the detection of trichome browning when flowers were sprayed with an alkaline chemical treatment.
Results
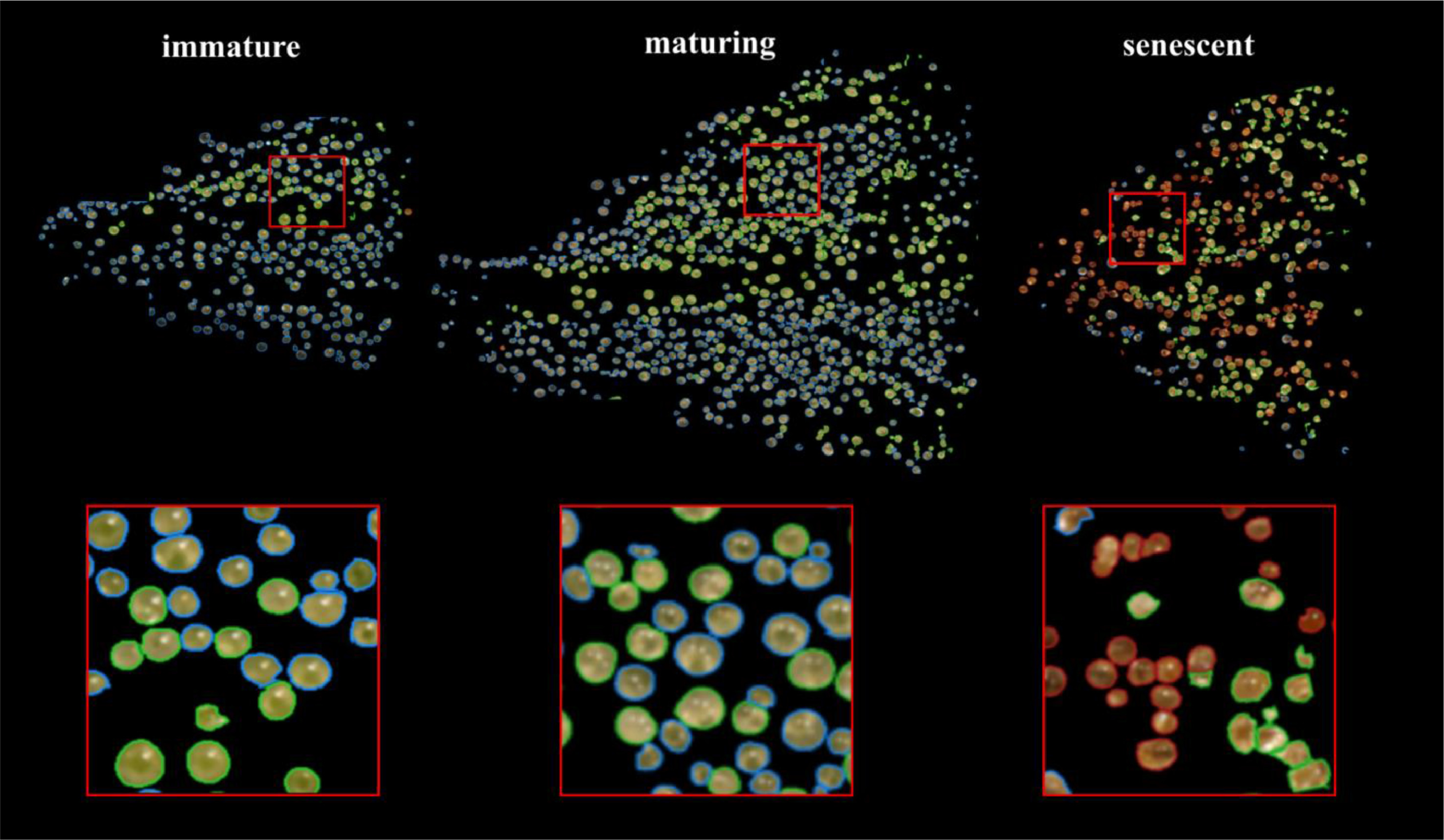
We manually inspected the accuracy of the predicted segmentation masks on a withheld test set of bract images. We observed a low false positive rate and satisfactory instance segmentation of whole trichome gland heads (Fig. 6). Glands that detached from stalks and released their contents to form amorphous blobs of resin were not detected. The accuracy of the k-NN classifier was verified via manual assessment of bract tissue images at different flowering time points to confirm trichome head phenotype predictions correctly described trichome gland development from transparent sessile and pre-stalked capitate (clear) to capitate stalked abundance (milky) prior to harvest time until trichome gland senescence (brown).
|
An examination of the clear-milky-brown results of all four cannabis strains over flowering time showed that in general, trichome browning was absent until approximately 50 days after flowering (daf) (Fig. 7). There was a rapid increase in the prevalence of brown trichome heads at 65–75 days of the flowering period. By comparison, the clear trichome heads were most abundant at day 25 of the flowering period, and began to steadily decline up to the end of the experiment (harvest time, day 65–75 of the flowering period, depending on the strain). The trichome heads displaying the milky phenotype began to gradually increase in prevalence in each strain, with a peak at day 55 in PK, at day 50 in strains AK and GDB, and a maximum at day 70 in WR. A significant difference in trichome gland head phenotype was not observed when assessed by dissection morphology, as outlined in Fig. 3.
|
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added. No other changes were made in accordance with the "NoDerivatives" portion of the license.