Difference between revisions of "Journal:Potential of NIRS technology for the determination of cannabinoid content in industrial hemp (Cannabis sativa L.)"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 379: | Line 379: | ||
Interest in the determination of the cannabinoid content of ''Cannabis sativa'' has increased in recent years, as indicated by the various studies published in this field. As in this study, other authors have achieved good results for the prediction of THC and CBD by NIRS without any data pretreatment. For example, Sánchez-Carnerero ''et al.'' [6] obtained an R<sup>2</sup>cv of 0.99 for CBD and an R<sup>2</sup>cv of 0.99 for THC with an FT-NIR instrument. However, using a NIR Systems 6500 scanning monochromator for CBD prediction, a SNV-DT pretreatment was applied, and resulted in an R<sup>2</sup>cv of 0.99 and a MSC for THC with an R<sup>2</sup>cv 0.98 and an RPD of 3.07. | Interest in the determination of the cannabinoid content of ''Cannabis sativa'' has increased in recent years, as indicated by the various studies published in this field. As in this study, other authors have achieved good results for the prediction of THC and CBD by NIRS without any data pretreatment. For example, Sánchez-Carnerero ''et al.'' [6] obtained an R<sup>2</sup>cv of 0.99 for CBD and an R<sup>2</sup>cv of 0.99 for THC with an FT-NIR instrument. However, using a NIR Systems 6500 scanning monochromator for CBD prediction, a SNV-DT pretreatment was applied, and resulted in an R<sup>2</sup>cv of 0.99 and a MSC for THC with an R<sup>2</sup>cv 0.98 and an RPD of 3.07. | ||
In a more recent study, Deidda ''et al.'' [37] explored the feasibility of using NIRS for the quantitative analysis of THC. For this study, two handheld NIR spectrophotometers were used and compared, a low-cost device (NIR-S-G1) and a mid-cost device (MicroNIR onsite W 1700). Both entire inflorescence and resin samples were analyzed, and the reference method used was UHPLC coupled to UV detection. A preliminary study was conducted on 26 entire inflorescences that were then ground and sieved in order to evaluate the impact of sample homogeneity on the THC content predictions. Researchers obtained a THC concentration far wider than in our study, ranging from 0.92 to 22.21%. RPD values between 1 and 4.54 were obtained for the different physical forms of samples using both devices. In general, the MicroNIR spectrophotometer outperformed NIR-S-G1. Moreover, 45 resin samples were analyzed with both devices, obtaining an RPD value of 2.26 with the MicroNIR and 1.51 with NIR-S-G1. Therefore, authors concluded that the mid-cost system was the best-suited spectrophotometer for their application. | |||
In another study carried out by Chen ''et al.'' [38], a different approach was adopted in which the authors explored the potential of NIRS for the ''in situ'' determination of CBD in hemp oil. For their study, 20 hemp oil samples with different concentrations of CBD and CBDA (determined by HPLC) were analyzed by a Bruker MATRIX-F FT-NIR spectrometer covering the 4000 to 12,000 cm<sup>−1</sup> range. Super partial least-squares regression (sPLSR) and a self-optimizing support vector elastic net (SOSVEN) were applied to predict the concentrations of CBD, achieving promising results with a coefficient of determination for the validation set >0.98 and an RMSEV of 6.4 ± 0.1 mg/mL. | |||
In addition, as mentioned in the introduction, spectroscopic techniques have been used to differentiate fiber-type from drug-type ''Cannabis sativa'' L. [39] In that study, ATR-FTIR in the 5000–400 cm<sup>−1</sup> region was used to assess 36 samples of ''C. sativa'' inflorescences; eight were drug-type, 14 fiber-type, and another set of 14 were cannabis samples having a low THC concentration. PLS models were developed to predict the content of seven neutral and acidic cannabinoids (THC, THCA, CBD, CBDA, CBG, [[cannabigerolic acid]] [CBGA], and [[cannabinol]] [CBN]). Authors achieved very good results, with an R<sup>2</sup>cv higher than 0.99 for each cannabinoid and RMSECV values ranging from 0.020 to 0.163. | |||
Moreover, mid-infrared (MIR) spectroscopy has been explored for the prediction of THC and CBD content in ''C. sativa''. For example, Geskovski ''et al.'' [40] used an ATR-FTIR spectrometer, in the 1700 to 400 cm<sup>−1</sup> range, to quantify the content of THC and CBD in 45 flowers and 34 cannabis extracts. PLS models were developed for both types of sample, obtaining good results with R<sup>2</sup>p and RMSEP values of 0.99% and 2.32% for THC, and 0.99% and 1.33% for CBD, respectively, for the flower samples, and R<sup>2</sup>p and RMSEP values of 0.95% and 3.79% for THC, and 0.99% and 1.44% for CBD, in the cannabis extract samples, respectively. | |||
In the current study, we present a feasible and low-cost method for THC and CBD content determination in ''C. sativa'' samples using NIRS. Promising results were obtained for both cannabinoids, indicating the potential of NIR technology as a predictive tool. | |||
==Conclusions== | |||
The functionality of NIRS for the quantification of THC and CBD as principal cannabinoids in hemp, along with their related spectral peaks, was evaluated in this study. According to the latter, the region of the NIR spectrum analyzed presents characteristic absorption bands around 1210, 1450, 1736, 1762, 1820, 1940, 2060, and 2090 nm that are typical of proteins, lipids, water, and other compounds present in hemp (from OH, NH, CH, and other bonds). In addition, the band at 1736 nm, related to aromatic hydrocarbons of the terpenes, was associated with the CBD content, since cannabinoids are terpene-phenolic compounds. | |||
Moreover, predictive models of the cannabinoid content in hemp were obtained combining NIR spectroscopy and chemometric analysis. The best results for the prediction of both THC and CBD were obtained using the raw data, providing a simpler form of analysis. For the THC, the best PLS model achieved a determination coefficient of cross-validation of 0.77 and an RPD value > 2, which indicates its predictive capacity. For the CBD, the best PLS model achieved a coefficient of 0.77 and an RPD value > 2, also indicating the goodness of the prediction model. | |||
Although the number of samples in this study was limited due to the high cost of HPLC, it allowed us to demonstrate the potential of NIRS for the determination of the main cannabinoid content in samples of the Kompolti variety. Due to the goodness of the models and the results obtained, this study may be extended to include a larger number of samples or other varieties of industrial hemp with a wider concentration of THC. | |||
The results obtained here demonstrate that NIR spectroscopy offers speed and simplicity unmatched by other traditional techniques. Accordingly, it was tested as an alternative to conventional HPLC analysis for the evaluation of cannabinoid content with promising results. | |||
==Acknowledgements== | |||
The authors would like to thank Genscore Navarra S.L. for access to the plant material samples used in this study. | |||
===Author contributions=== | |||
Conceptualization, C.J., A.Á. and S.A.; Data curation, S.A., A.L.-M. and C.J.; Formal analysis, S.A., C.P.-R., A.L.-M. and P.C.Z.; Funding acquisition, C.J. and A.Á.; Investigation, C.P.-R., A.L.-M., C.J., P.C.Z. and S.A.; Methodology, P.C.Z., C.J., C.P.-R., A.L.-M. and S.A.; Project administration, A.Á. and S.A.; Resources, C.J., A.Á. and S.A.; Software, S.A.; Supervision, C.P.-R., A.L.-M., C.J. and S.A.; Validation, P.C.Z., C.P.-R., A.L.-M. and C.J.; Visualization, C.P.-R. and A.L.-M.; Writing—original draft, P.C.Z., C.J. and A.L.-M.; Writing—review and editing, P.C.Z., C.P.-R., A.L.-M., C.J., A.Á. and S.A. All authors have read and agreed to the published version of the manuscript. | |||
===Funding=== | |||
This research received no external funding. Open access co-funding provided by Institute for Sustainability & Food Chain Innovation (IS-FOOD). | |||
===Conflicts of interest=== | |||
The authors declare no conflict of interest. | |||
==References== | ==References== | ||
Revision as of 16:46, 11 May 2022
| Full article title | Potential of NIRS technology for the determination of cannabinoid content in industrial hemp (Cannabis sativa L.) |
|---|---|
| Journal | Agronomy |
| Author(s) | Jarén, Carmen; Zambrana, Paula C.; Pérez-Roncal, Claudia; López-Maestresalas, Ainara; Ábrego, Andrés; Arazuri, Silvia |
| Author affiliation(s) | Universidad Pública de Navarra, Genscore Navarra S.L. |
| Primary contact | Email: cjaren at unavarra dot es |
| Year published | 2022 |
| Volume and issue | 12(4) |
| Article # | 938 |
| DOI | 10.3390/agronomy12040938 |
| ISSN | 2073-4395 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.mdpi.com/2073-4395/12/4/938/htm |
| Download | https://www.mdpi.com/2073-4395/12/4/938/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Industrial hemp (Cannabis sativa L.) is a plant native to Asia and is considered to be a primary source of food, textile fiber, and medicines. It is characterized by containing minimal concentrations of Δ9-tetrahydrocannabinol (THC), which is the main psychoactive chemical component, and cannabidiol (CBD), a non-psychoactive substance. In most European countries, the maximum concentration legally allowed for cultivation is 0.2% of THC, and it is currently under debate whether to increase this level to 0.3%. Moreover, in many countries its production is being regulated and legalized, increasing the need for a rapid analysis method.
The present work evaluated the cannabinoid content in hemp using near-infrared spectroscopy (NIRS) technology in combination with chemometric techniques. For this, several samples of the Kompolti variety were analyzed. Samples were dried and ground, and the content of total THC (%) and total CBD (%) was determined by high-performance liquid chromatography (HPLC) with a diode array detector for reference measurements, and then the spectra were collected by NIRS. Principal component analysis and partial least square regression models were developed. Good coefficients of determination of cross-validation of 0.77 for THC and CBD, and a ratio of prediction to deviation >2 for total THC and CBD, were achieved. The results obtained show that NIRS technology has potential for the quantitative determination of cannabinoids. Therefore, this analytical method would allow a simpler, more robust, precise, and sustainable estimation than the current HPLC approach.
Keywords: CBD, THC, near infrared spectroscopy, quantification, HPLC, chemometrics
Introduction
The non-psychotropic species Cannabis sativa L., referred to as industrial hemp [1], is characterized by containing minimal concentrations of Δ9-tetrahydrocannabinol (THC), the main psychoactive chemical component, and cannabidiol (CBD), a non-psychoactive substance that is often present in amounts similar to those of THC. [1,2] Hemp is mainly used for food or textile purposes and, in addition, offers great medicinal potential. Although the regulations of different countries vary according to the definition of the maximum accepted THC limit, industrial-hemp-producing countries require that the varieties used contain THC concentrations lower than 1%. In most European countries, the current upper legal limit for cultivation is 0.2% of THC and the ratio of CBD to THC should be greater than one. Currently, the maximum concentration legally permitted for cultivation is under debate in the European Union. [3]
It is important to note that the flower is the part of hemp with the highest significant content of cannabinoids. [4] These, when heated, spontaneously decarboxylate to the “neutral” cannabinoids THC and CBD. This heat-labile characteristic of acidic cannabinoids (e.g., tetrahydrocannabinolic acid [THCA] and cannabidiolic acid [CBDA]) highlights the importance of using a low-temperature, non-destructive method to achieve a precise quantification of these molecules. [5] Moreover, for all stakeholders in the cannabis supply chain, a precise and trustworthy identification of these cannabinoids would be of great economic importance. [4]
Traditionally, cannabinoid content has been determined by high-performance liquid chromatography (HPLC) and gas chromatography (GC). HPLC provides a full cannabinoid profile, but it has several associated disadvantages, including sample destruction, complex instrumentation, involvement of hazardous chemicals, and longer sample preparation times, which limit its application on-site, where a fast and non-destructive process is preferable. [4] Similarly, GC is another preferred method for the determination of these compounds. However, it is a slow and expensive technique, requiring a tedious sample preparation stage that involves the extraction of the active ingredients through the use of organic solvents, whose subsequent residues must be managed with a considerable increase in cost and time. [6]
These limitations have led to a search for faster and easier-to-use alternatives to HPLC and GC. [4] Therefore, it is important to develop a simple, fast, and sustainable method for the quantification of cannabinoids. In recent years, spectroscopic methods have emerged as techniques that are used on a wide range of biological samples without the need for extraction. [7] One such technique is near-infrared spectroscopy (NIRS), which is a fast, cost-effective, versatile, robust, and sustainable technique. In addition, it allows both quantitative and qualitative determinations of the main parameters, such as proteins, fats, humidity, ashes, starch, or sugar, of the raw materials related to the quality of agricultural products. [6] In recent years, the interest in NIRS applied to hemp has gained importance due to the moisture, volatile substances, and chemical compounds in herbal products absorbed in the NIR region. In general, NIRS combined with chemometrics has great potential in the analysis of natural plant products. [8]
It should be taken into account that the cannabis flower is heterogeneous in nature, which presents a series of problems and drawbacks. It is a complex matrix, made up of a great variety of types of plant tissues and more than 500 different naturally produced chemicals. Moreover, it is a material that can vary widely between plants of the same crop, in an individual plant, and even within the same sample [9]. Consequently, no two parts of the cannabis flower are alike, and their cannabinoid content is likely to vary widely. In this scenario, NIRS technology is an adequate alternative for the analysis of heterogeneous vegetal samples and may therefore overcome the inherent heterogeneity of the cannabis plant. [4]
NIRS has been applied to discriminate between cannabis “drug type” (chemotype I) and “fiber type” (chemotype II) [10], for the discrimination of leaves of Cannabis sativa L. and other plant species [11], and for the prediction of the growth stage of cannabis plants in the early stages of cultivation. [12]
Marcel et al. [13] developed a prediction model of the chemical composition of the fiber and the central fraction of hemp (chemotype III) using NIRS combined with a partial least squares (PLS) regression analysis. Similarly, a procedure was developed for the identification and quantitative determination of synthetic cannabinoids in illicit herbal samples. The methodology was based on the measurement by Fourier-transform infrared spectroscopy of attenuated total reflectance (ATR-FTIR). [14]
Moreover, the total content of THC and CBD in the cannabis flower has been determined by Fourier-transform near-infrared spectroscopy (FT–NIR). [4] Similarly, Sánchez-Carnerero et al. [6] studied the prediction of cannabinoid content using NIRS. They used both FT-NIR and NIR spectrophotometers for their analysis and compared the results obtained with the two techniques. Similar results were obtained using both instruments, thus confirming that there is enough information in the spectral region of the NIR for the prediction of cannabinoids.
More recently, Duchateau et al. [15] created two classification methods according to the European laws about the discrimination of the legal limits of Cannabis spp. using NIR. Valinger et al. [7] described the development of artificial neural network (ANN) models for the prediction of the physical and chemical properties of industrial hemp extracts, based on the combination of ultraviolet-visible near-infrared (UV-VIS-NIR) spectra. For this, two different extraction methods were prepared (solid–liquid extraction and microwave-assisted extraction). The results showed that reliable ANN models can be developed to describe the physical and chemical characteristics, without the need for pre-processing of the spectra. In a recent study, Risoluti et al. [16], using a MicroNIR spectrometer, developed a test for cannabinoid determination in commercial hemp flours spiked with THC, CBD, and cannabigerol (CBG).
Therefore, the aim of this study was to evaluate the functionality of NIRS for the quantification of the main cannabinoids present in hemp samples. In addition, a study of the NIR spectra was carried out to identify the peaks.
Materials and methods
Vegetal material
Thirty-five hemp samples were obtained in collaboration with Genscore Navarra S.L. The specimens obtained were of the Kompolti variety, which is among the varieties authorized for the cultivation of industrial hemp in Spain. [17]
The plant material was weighed on a AB104 Mettler-Toledo analytical balance and dried in an oven at 60 °C for 24 hours, until a humidity between 8% and 13% was achieved, as recommended by regulation (EU) 2017/1155. [18]
Stems and seeds of more than 2 mm were removed from the dry samples and, with the help of a mortar, they were crushed until obtaining a semi-fine powder, in such a way that it could pass through a 1 mm mesh sieve. The samples were stored, without crushing them, in a dark place at a temperature below 25 °C. [18,19]
Spectra acquisition
Spectra were collected using an Acousto-Optic Tunable Filter (AOTF) NIR and Indium Gallium Arsenide (InGaAs) detector, called a Luminar 5030 Miniature "Hand-held," in the reflectance mode and equipped with Snap32! software (Brimrose Corporation of America, Sparks, MD, USA). A spectral range of 1200–2200 nm was used to obtain the spectra, with a sampling interval of 2 nm, and scanning speed of 60 ms. Each spectrum recorded by the instrument was the average of 50 scans.
In this study, 3 g of each of the 35 hemp powder samples was weighed and placed on the rotating cell of the AOTF-NIR spectrophotometer. As the sample rotates, the spectrum is measured so that different parts of the sample are scanned from above and inhomogeneities averaged. For each individual sample, three reflectance spectra were acquired by contacting the probe with the sample.
Reference measurements: HPLC
After acquisition of the NIRS data, the same hemp samples were removed from the Petri dishes and sent to a certified laboratory, ANANDA ANALYTICS LAB S.L., where an HPLC with diode array detector (HPLC-DAD) method was used for the determination of total THC and total CBD cannabinoids. The extraction was performed by ultrasound with subsequent methanol-chloroform decarboxylation. The mobile phase was acetonitrile (water [8:2 v/v], isocratic, stop time 8 minutes) according to Recommended methods for the identification and analysis of cannabis and cannabis products by the United Nations Office on Drugs and Crime. [19] Regarding the result, for a qualitative identification, the retention time and the DAD spectrum of the cannabinoid must match. The calculation for the quantitative results was carried out at the wavelengths of 220 and 240 nm.
The results were received after 10 days and, then, these reference chromatographic data were correlated with the spectral information to generate the NIR models for total THC and CBD prediction. Thirty-two of 35 samples were analyzed by HPLC-DAD, with three fungus-infected hemp plants being identified and excluded.
Multivariate data analysis
Data analysis was performed using the specific software The Unscrambler X v.10.4 (Camo Software AS, Oslo, Norway). First, principal component analysis (PCA) was performed with the full set of samples. PCA was applied to explore the spectral variability of the population [20], which also allows elimination of outliers that can have a negative effect on modeling. [21]
During the pre-model building phase, data pre-processing was carried out to eliminate the non-informative effects of light scattering or system noise. For the development of the models, in addition to working with the raw data, different data pretreatments were applied: spectra normalization, standard normal variate (SNV), standard normal variate and detrend (SNV-DT), multiplicative scatter correction (MSC), and first derivative. These are described as such:
- Raw data: The absolute reflectance was obtained from the radiation measurements of the 35 samples, with three repetitions each.
- Spectra normalization: Raw NIR spectra are often mathematically processed prior to development of the calibration model; such treatments include normalization, which is performed to minimize unwanted sources of data variation prior to calibration and to improve spectral characteristics. [22] Mean normalization was performed in this study, which is the most classical approach. In this normalization, each sample of the dataset (each row of the data matrix) is divided by its average.
- SNV: To eliminate interferences due to path length effects, SNV consists of subtracting the mean value of the spectrum from each reflectance value at each wavelength and dividing it by the standard deviation. [23]
- SNV-DT: This method was developed by Barnes et al. [24] to eliminate multiplicative scattering interferences and particle size, and to take into account the variation in the baseline change and curvilinearity in diffuse reflectance spectra. Detrend consists of fitting a second-order polynomial to the spectrum corrected by SNV, which is subtracted to eliminate the dispersion effect that is dependent on each wavelength. [25]
- MSC: This is another preprocessing technique that corrects the displacements between samples due to the particles of the samples. [26] MSC is undertaken by using a reference spectrum and correcting the different spectra to it so that the baseline and the amplification effects are at the same average level in all spectra. [27] The basic concept of MSC is to remove non-linearities in the data caused by scattering from particulates in the samples. [28]
- First derivative: The first derivative of the spectra, based on the Savitzky–Golay algorithm, is used to increase the spectral resolution and interpret the spectra. [29] One smoothing point was applied to the right and another to the left, and a polynomial of order two was used to smooth and eliminate random noise from the NIR spectra. Vasques et al. [30] confirmed that the Savitzky–Golay derivatives were among the best methods for preprocessing the spectra. Similarly, Ertlen et al. [31] reported that by using derivatives, more convenient information can be taken from NIR spectra.
After data pretreatments, a PLS regression analysis was applied to the dataset to build a model capable of predicting the content of cannabinoids, both for total THC and total CBD, in the hemp samples, and to be able to assess the effectiveness of NIR spectroscopy. For the validation of the model, cross-validation (CV) was used in order to calculate the relationships between spectral and chemical properties. Williams et al. [32] recommend CV for the evaluation of any calibration model based on small sets of samples below 100 units.
The performance of the calibration models was evaluated using the root mean square error of the calibration (RMSEC), the root mean square error of cross-validation (RMSECV), the coefficient of determination of calibration (R2c), the coefficient of determination of cross-validation (R2cv), and the ratio of prediction to deviation (RPD). The number of latent variables (LV) was used to prevent overfitting.
Results and discussions
HPLC results
For each of the samples analyzed, an individualized report was received with the sample identification data, the characteristics of the analysis methods used, and the results obtained from the HPLC-DAD analysis. The results provided by the reference laboratory, expressed as a percentage, included data for humidity, THC total, CBD total, and 12 other cannabinoids that were not the subject of this study. Thirty-two samples of 35 were analyzed by HPLC-DAD because three hemp plants infected with fungi were identified. The mean, the maximum, the minimum, and the standard deviation of the reference results can be seen in Table 1.
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Regarding the percentage of humidity, all samples presented values between 9.02% and 12.34%. These values are between the ranges established in the delegated regulation [18], which indicates a humidity of 8–13% in the preparation of cannabis samples. All selected samples had a THC value detected between 0.057% and 0.161%. Concerning CBD, which is currently the compound of greatest interest, the results in Table 1 show a maximum value of 5.342% and a minimum value of 2.178%, which indicates the high concentration of CBD in the samples, a characteristic of industrial cannabis. [19]
Spectral data
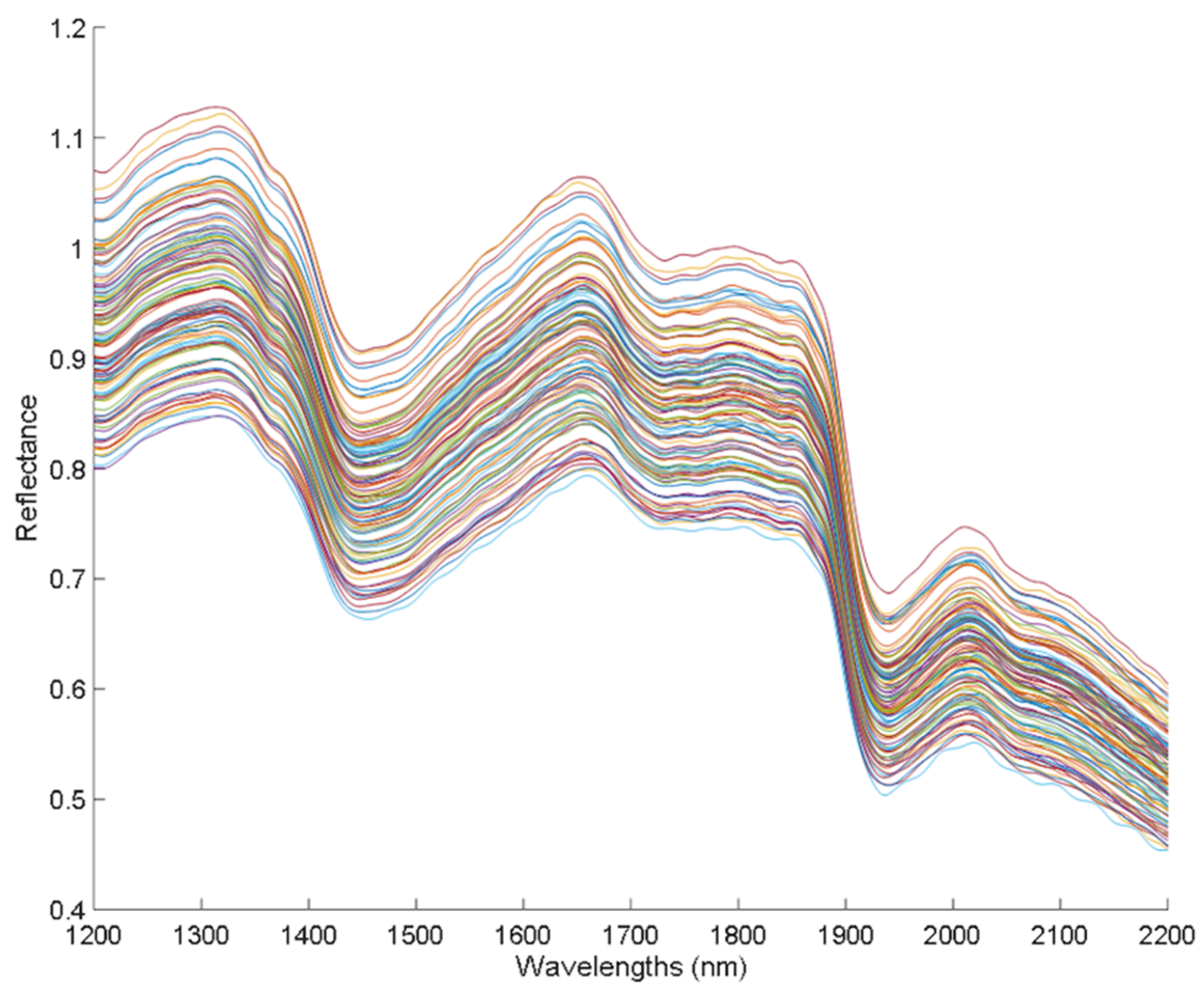
In Figure 1, the spectra of the hemp sample set are shown, which correspond to the spectra collected in the Luminar 5030 AOTF-NIR spectrophotometer, where the typical reflectance bands of hemp appear in the NIR region from 1200 to 2200 nm.
|
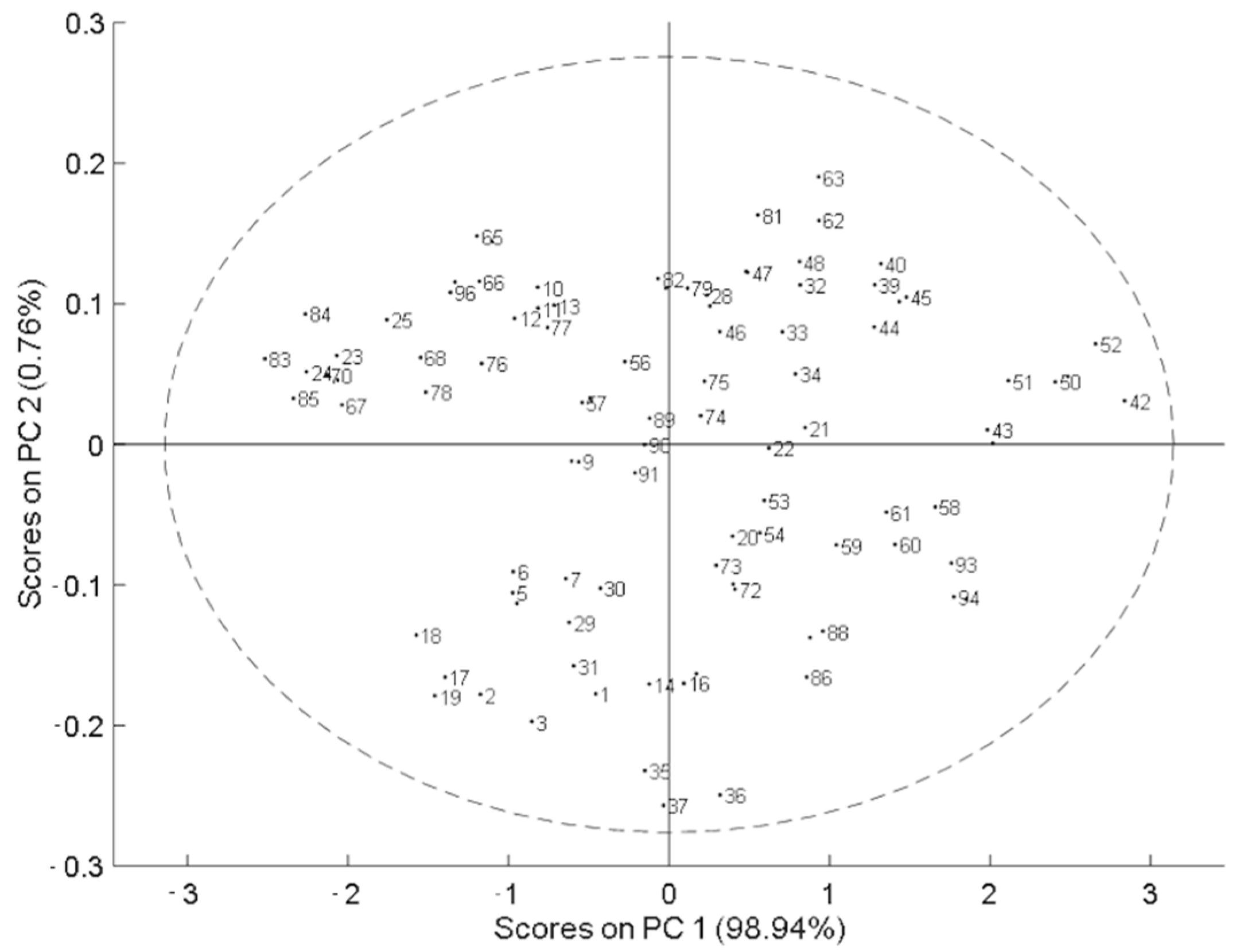
Principal component analysis (PCA) was applied to explore the spectral variability of the population, where no atypical samples were identified. This indicates that the dataset was uniformly distributed and covered as much spectral variation as possible (Figure 2). Then, PLS analysis was applied to the dataset. The results for each PLS model, for total THC and total CBD, with each pretreatment applied, including the number of samples used, were compared based on the values obtained for RMSEC, RMSECV, R2c, R2cv, LV, SD, and RPD.
|
Interpretation of spectra
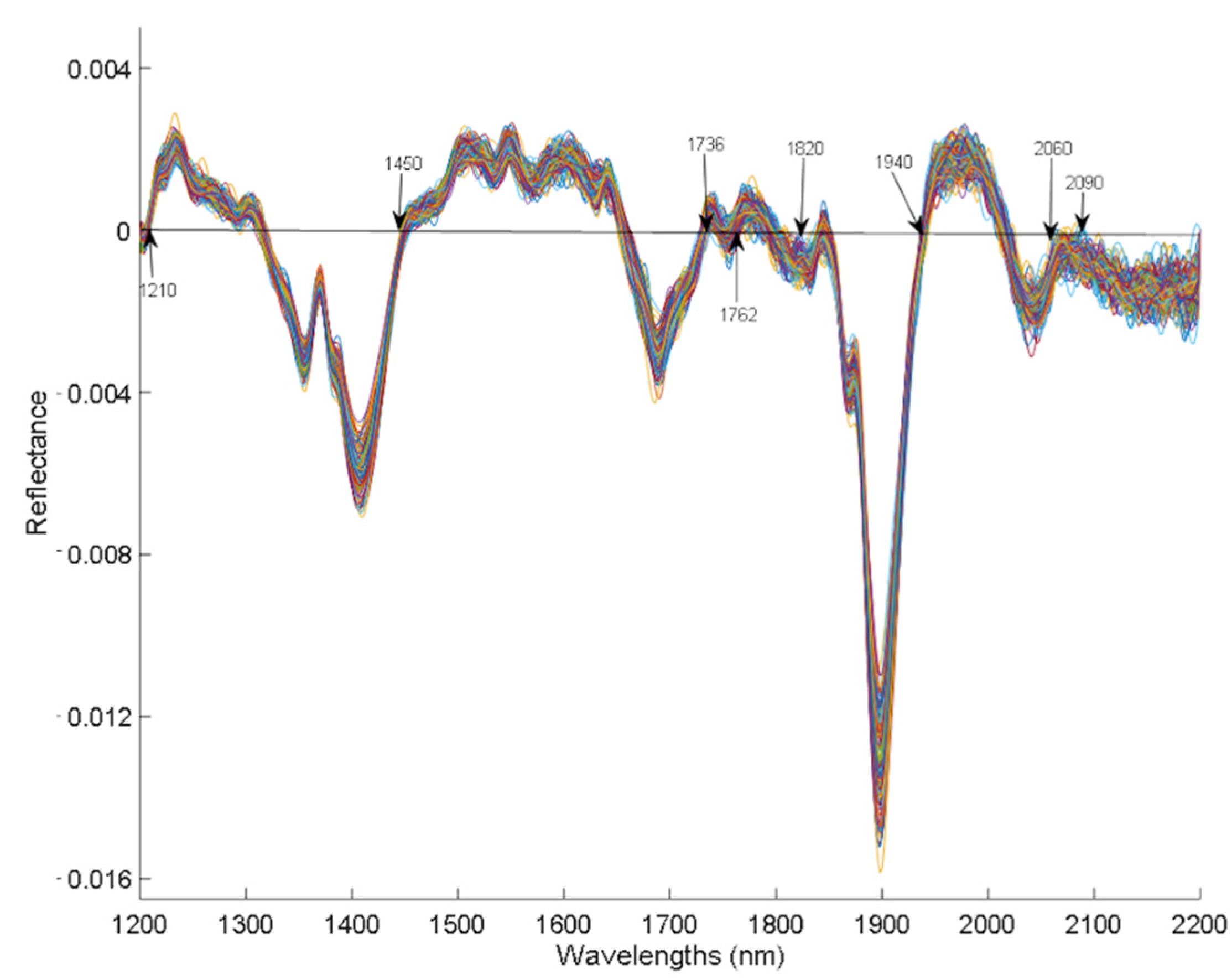
Figure 3 shows the reflectance spectra of the samples after applying the first derivative based on the Savitzky–Golay algorithm. The typical absorption bands of plant material appear in the NIR region derived from the superpositions corresponding mainly to overtones and to the combination of vibration modes that involve chemical bonds of the types C-H, O-H, and N-H. [8] In NIR reflectance spectra, the absorption bands of the molecules that make up the sample are described as valleys rather than peaks, since the absorbance equals Log (1/Reflectance). The hemp spectra obtained in this study show the characteristic bands around 1210, 1450, 1736, 1762, 1820, 1940, 2060, and 2090 nm that are characteristic of proteins, lipids, water, and other compounds present in hemp in the main absorptions (of OH, NH, CH, and other bonds).
|
The 1450 and 1940 nm bands that appear in the spectra correspond to the first overtone and to the -OH group combination bands, mainly due to the presence of water. Although the samples were oven dried at 60 °C, they were left with a humidity between 8 and 13%, which may be responsible of these absorption bands. Bands associated with lipids were found around 1210 nm (second overtone of stretching vibrations) and 1762 nm (first overtone of stretching vibrations) of functional groups -CH and -CH2 corresponding to aliphatic hydrocarbons. Bands around 2060 nm have been associated with protein absorption, which are related to NH secondary combination bands of amides in proteins, for peptide groups, and those around 1820 and 2090 nm correspond to associated bands of cellulose and polymeric structure, which are related to the fiber content of hemp. [33,34] In addition, the reflectance band found at 1736 nm corresponds to the aromatic hydrocarbons of the terpenes, of the functional group C-H. [33,34,35] This band is more evident in samples that have a higher CBD content, because terpenes and cannabinoids share biosynthetic pathways, and because cannabinoids are terpene-phenolic compounds. [6] In general, the absorption bands obtained in the present study are similar to those reported by other similar studies on cannabinoids. [6]
PLS model for total THC
PLS analysis was applied to the dataset using the same number of samples for both calibration and CV predictive models. The results of each PLS model for total THC applied with each pretreatment include the number of samples used, RMSEC, RMSECV, R2c, R2cv, SD, and RPD.
The results for the R2cv of the predictive models were higher than 0.72, which indicates that the calibration models can be considered good. [20]
Similarly, the RPD was considered a good indicator of the predictive capacity. In the prediction of total THC content, RPD values > 2 were obtained, which indicates the goodness of the models. The higher the RPD value, the greater the ability of the calibration model to accurately predict the reference parameter values (Table 2).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Some authors have established five levels of prediction accuracy based on the RPD. Saeys et al. [36] indicated that models with RPD values below 1.5 should not be used for prediction; an RPD value between 1.5 and 2.0 should only be used to distinguish between high and low values; and a value between 2.0 and 2.5 can be used to make approximate quantitative predictions. Values between 2.5 and 3.0 and above 3.0 can be used for good and excellent predictions, respectively.
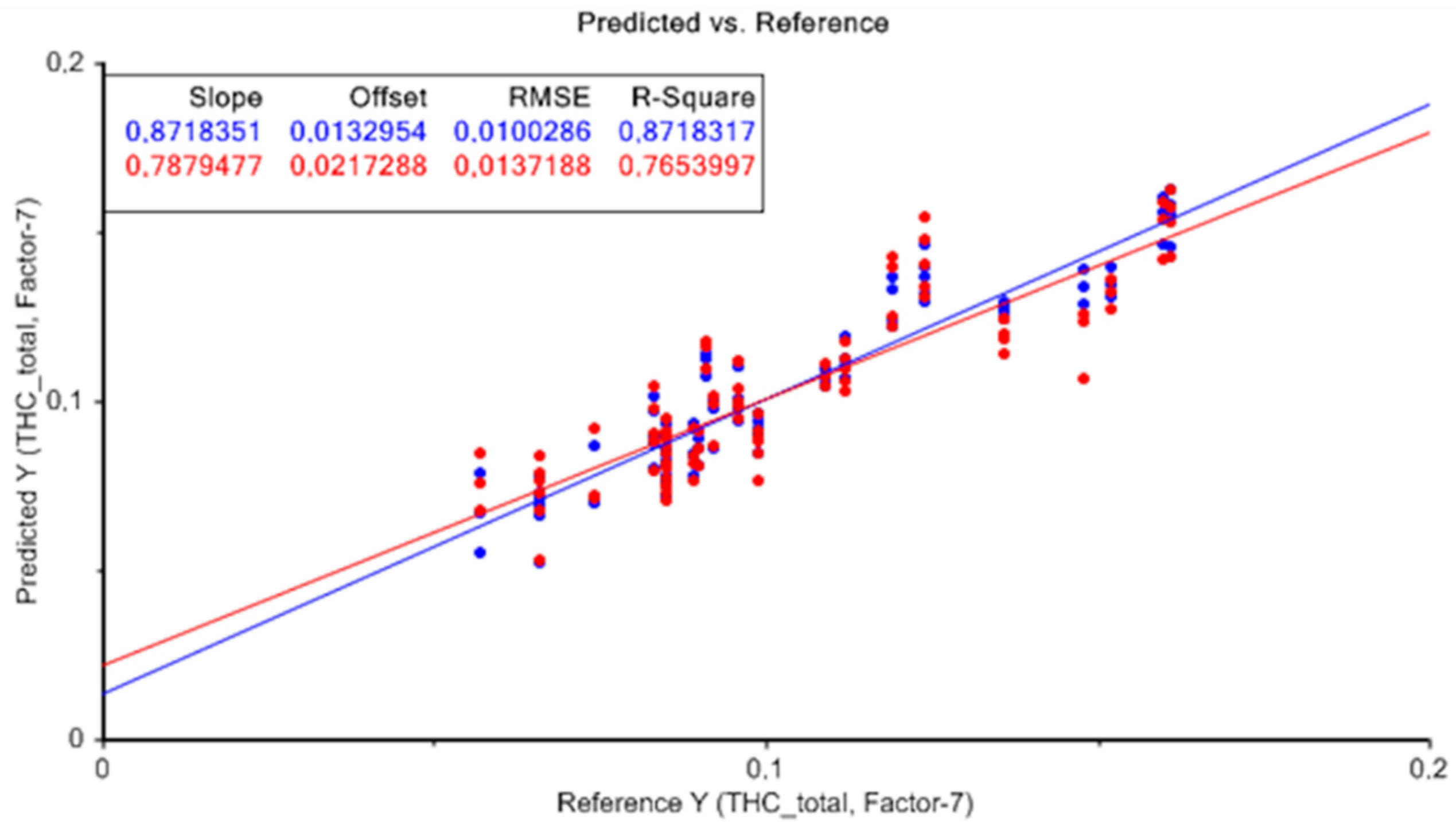
For total THC (%), the best model was selected according to the highest values of R2cv and RPD, and the lowest values of RMSECV. The results without applying any data pretreatment technique presented higher values of R2c of 0.87, R2cv of 0.77, and an RPD > 2, which indicates that approximate quantitative predictions are possible. The lower RMSECV values of 0.014 were also taken into account.
Figure 4 represents the result of the PLS model without applying any data pretreatment technique, for the quantification of total THC. The graph shows measured values (X-axis) versus predicted values (Y-axis) for total THC from the cross-validation dataset showing analogous results. The proximity between the calibration line and the cross-validation line can be observed with slopes very close to the unit, which provides an idea of the quality of the model obtained.
|
In general, the results showed that the developed models had the ability to make approximate predictions of total THC content, with high R2 and RPD, and low RMSE. Furthermore, the difference between RMSEC and RMSECV was low in all models developed. Therefore, for the determination of total THC, the prediction model obtained without applying any data pretreatment technique presented the best results, taking seven latent variables. For the rest of the calibration models carried out applying different combinations of data pretreatments, similar results were obtained with minimal differences.
PLS model for total CBD
The results for the R2cv of the predictive models were higher than 0.73, which indicates that the calibration models can be considered good, except for the first derivative model, which has an R2cv value of 0.68. The latter would allow adequate discrimination between samples having high, medium, and low total CBD content. [20]
The standard deviation of CBD content was equal to 0.892, considering only the samples used for each calibration model, and 0.871 for the first derivative model. In general, the results obtained for the RPD were around 2, which indicates the possibility for approximate quantitative predictions (Table 3).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
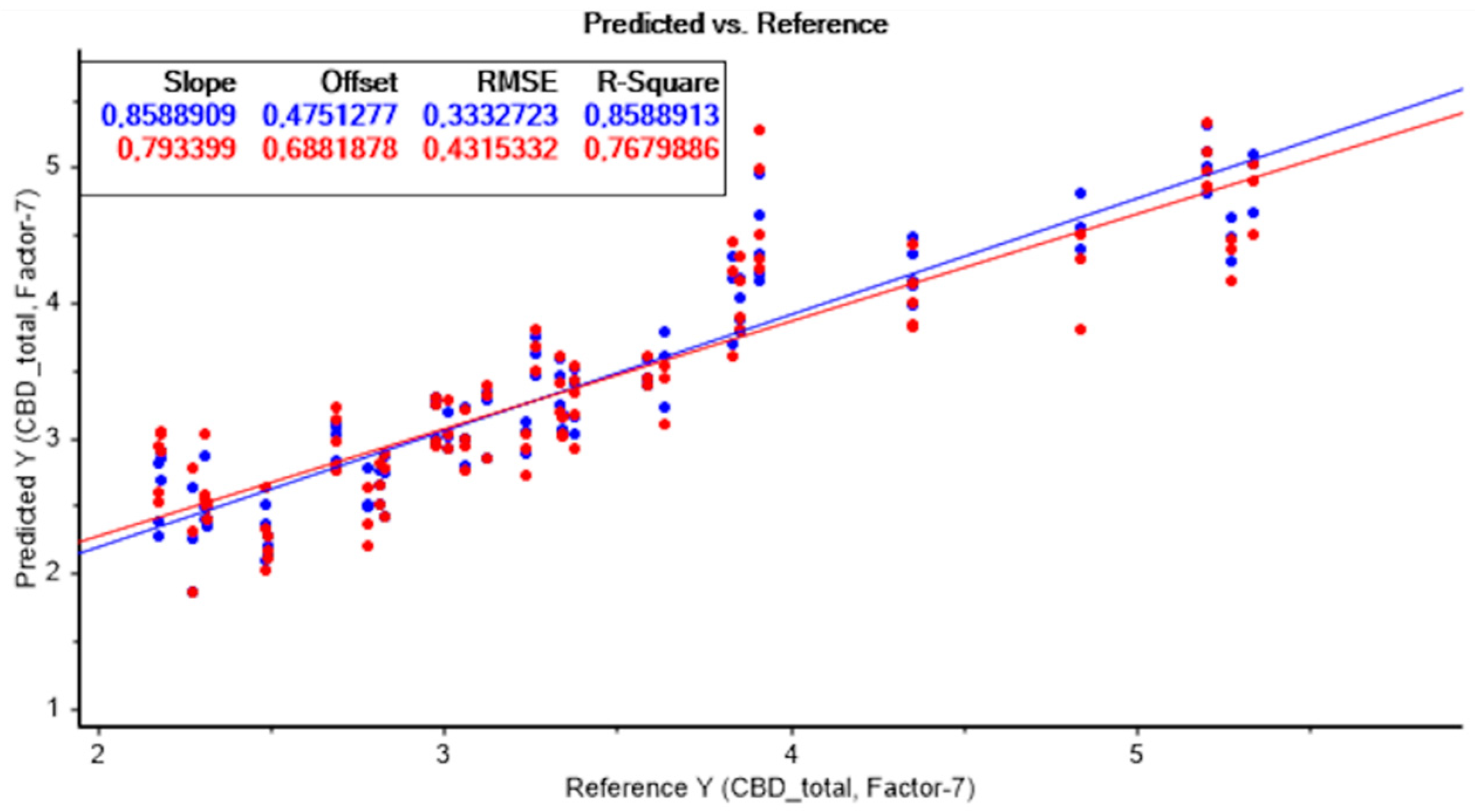
The best model to predict CBD content was also obtained without applying any data pretreatment technique. A high R2c value of 0.86, R2cv of 0.77, and an RPD > 2 were achieved, indicating the predictive capacity of the model. The lower RMSECV values of 0.431 were also considered.
Figure 5 represents the result of the PLS model without applying any data pretreatment technique, for the quantification of total CBD.
|
Moreover, as with the prediction of THC, all developed models showed an excellent performance in predicting total CBD content, with high R2 and RPD, and low RMSE, which demonstrates the potential of NIRS for cannabinoid content estimation.
Furthermore, the difference between RMSEC and RMSECV was low in all models developed. Therefore, for the determination of the total CBD, the prediction model obtained without applying any data pre-treatment technique presented the best results, taking seven latent variables. Comparable results were obtained for the rest of the calibration models carried out applying different combinations of data pretreatments.
Interest in the determination of the cannabinoid content of Cannabis sativa has increased in recent years, as indicated by the various studies published in this field. As in this study, other authors have achieved good results for the prediction of THC and CBD by NIRS without any data pretreatment. For example, Sánchez-Carnerero et al. [6] obtained an R2cv of 0.99 for CBD and an R2cv of 0.99 for THC with an FT-NIR instrument. However, using a NIR Systems 6500 scanning monochromator for CBD prediction, a SNV-DT pretreatment was applied, and resulted in an R2cv of 0.99 and a MSC for THC with an R2cv 0.98 and an RPD of 3.07.
In a more recent study, Deidda et al. [37] explored the feasibility of using NIRS for the quantitative analysis of THC. For this study, two handheld NIR spectrophotometers were used and compared, a low-cost device (NIR-S-G1) and a mid-cost device (MicroNIR onsite W 1700). Both entire inflorescence and resin samples were analyzed, and the reference method used was UHPLC coupled to UV detection. A preliminary study was conducted on 26 entire inflorescences that were then ground and sieved in order to evaluate the impact of sample homogeneity on the THC content predictions. Researchers obtained a THC concentration far wider than in our study, ranging from 0.92 to 22.21%. RPD values between 1 and 4.54 were obtained for the different physical forms of samples using both devices. In general, the MicroNIR spectrophotometer outperformed NIR-S-G1. Moreover, 45 resin samples were analyzed with both devices, obtaining an RPD value of 2.26 with the MicroNIR and 1.51 with NIR-S-G1. Therefore, authors concluded that the mid-cost system was the best-suited spectrophotometer for their application.
In another study carried out by Chen et al. [38], a different approach was adopted in which the authors explored the potential of NIRS for the in situ determination of CBD in hemp oil. For their study, 20 hemp oil samples with different concentrations of CBD and CBDA (determined by HPLC) were analyzed by a Bruker MATRIX-F FT-NIR spectrometer covering the 4000 to 12,000 cm−1 range. Super partial least-squares regression (sPLSR) and a self-optimizing support vector elastic net (SOSVEN) were applied to predict the concentrations of CBD, achieving promising results with a coefficient of determination for the validation set >0.98 and an RMSEV of 6.4 ± 0.1 mg/mL.
In addition, as mentioned in the introduction, spectroscopic techniques have been used to differentiate fiber-type from drug-type Cannabis sativa L. [39] In that study, ATR-FTIR in the 5000–400 cm−1 region was used to assess 36 samples of C. sativa inflorescences; eight were drug-type, 14 fiber-type, and another set of 14 were cannabis samples having a low THC concentration. PLS models were developed to predict the content of seven neutral and acidic cannabinoids (THC, THCA, CBD, CBDA, CBG, cannabigerolic acid [CBGA], and cannabinol [CBN]). Authors achieved very good results, with an R2cv higher than 0.99 for each cannabinoid and RMSECV values ranging from 0.020 to 0.163.
Moreover, mid-infrared (MIR) spectroscopy has been explored for the prediction of THC and CBD content in C. sativa. For example, Geskovski et al. [40] used an ATR-FTIR spectrometer, in the 1700 to 400 cm−1 range, to quantify the content of THC and CBD in 45 flowers and 34 cannabis extracts. PLS models were developed for both types of sample, obtaining good results with R2p and RMSEP values of 0.99% and 2.32% for THC, and 0.99% and 1.33% for CBD, respectively, for the flower samples, and R2p and RMSEP values of 0.95% and 3.79% for THC, and 0.99% and 1.44% for CBD, in the cannabis extract samples, respectively.
In the current study, we present a feasible and low-cost method for THC and CBD content determination in C. sativa samples using NIRS. Promising results were obtained for both cannabinoids, indicating the potential of NIR technology as a predictive tool.
Conclusions
The functionality of NIRS for the quantification of THC and CBD as principal cannabinoids in hemp, along with their related spectral peaks, was evaluated in this study. According to the latter, the region of the NIR spectrum analyzed presents characteristic absorption bands around 1210, 1450, 1736, 1762, 1820, 1940, 2060, and 2090 nm that are typical of proteins, lipids, water, and other compounds present in hemp (from OH, NH, CH, and other bonds). In addition, the band at 1736 nm, related to aromatic hydrocarbons of the terpenes, was associated with the CBD content, since cannabinoids are terpene-phenolic compounds.
Moreover, predictive models of the cannabinoid content in hemp were obtained combining NIR spectroscopy and chemometric analysis. The best results for the prediction of both THC and CBD were obtained using the raw data, providing a simpler form of analysis. For the THC, the best PLS model achieved a determination coefficient of cross-validation of 0.77 and an RPD value > 2, which indicates its predictive capacity. For the CBD, the best PLS model achieved a coefficient of 0.77 and an RPD value > 2, also indicating the goodness of the prediction model.
Although the number of samples in this study was limited due to the high cost of HPLC, it allowed us to demonstrate the potential of NIRS for the determination of the main cannabinoid content in samples of the Kompolti variety. Due to the goodness of the models and the results obtained, this study may be extended to include a larger number of samples or other varieties of industrial hemp with a wider concentration of THC.
The results obtained here demonstrate that NIR spectroscopy offers speed and simplicity unmatched by other traditional techniques. Accordingly, it was tested as an alternative to conventional HPLC analysis for the evaluation of cannabinoid content with promising results.
Acknowledgements
The authors would like to thank Genscore Navarra S.L. for access to the plant material samples used in this study.
Author contributions
Conceptualization, C.J., A.Á. and S.A.; Data curation, S.A., A.L.-M. and C.J.; Formal analysis, S.A., C.P.-R., A.L.-M. and P.C.Z.; Funding acquisition, C.J. and A.Á.; Investigation, C.P.-R., A.L.-M., C.J., P.C.Z. and S.A.; Methodology, P.C.Z., C.J., C.P.-R., A.L.-M. and S.A.; Project administration, A.Á. and S.A.; Resources, C.J., A.Á. and S.A.; Software, S.A.; Supervision, C.P.-R., A.L.-M., C.J. and S.A.; Validation, P.C.Z., C.P.-R., A.L.-M. and C.J.; Visualization, C.P.-R. and A.L.-M.; Writing—original draft, P.C.Z., C.J. and A.L.-M.; Writing—review and editing, P.C.Z., C.P.-R., A.L.-M., C.J., A.Á. and S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. Open access co-funding provided by Institute for Sustainability & Food Chain Innovation (IS-FOOD).
Conflicts of interest
The authors declare no conflict of interest.
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added.