Journal:Cannabis sativa research trends, challenges, and new-age perspectives
| Full article title | Cannabis sativa research trends, challenges, and new-age perspectives |
|---|---|
| Journal | iScience |
| Author(s) | Hussain, Tajammul; Jeena, Ganga; Pitakbut, Thanet; Vasilev, Nikolay; Kayser, Oliver |
| Author affiliation(s) | TU Dortmund University |
| Primary contact | Email: Tajammul dot hussain at tu-dortmund dot de |
| Year published | 2021 |
| Volume and issue | 24(12) |
| Article # | 103391 |
| DOI | 10.1016/j.isci.2021.103391 |
| ISSN | 2589-0042 |
| Distribution license | Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International |
| Website | https://www.sciencedirect.com/science/article/pii/S2589004221013626 |
| Download | https://www.sciencedirect.com/science/article/pii/S2589004221013626/pdfft (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Background: Cannabis sativa L. is one of the oldest known medicinal plants, cultivated for at least 10,000 years for several agricultural and industrial applications. However, the plant became controversial owing to some psychoactive components that have adverse effects on human health.
Methods: In this review, we analyze the trends in cannabis research for the past two centuries. We discuss the historical transitions of cannabis from the category of "herbal medicine" to an illicit drug and back to a medicinal product post-legalization. In addition, we address the new-age application of immuno-suppressive and anti-inflammatory cannabis extracts for the treatment of COVID-19 inflammation. We further address the influence of the legal aspects of cannabis cultivation for medicinal, pharmaceutical, and biotechnological research. Finally, we review the up-to-date cannabis-related genomic resources and advanced technologies for their potential application in genomic-based cannabis improvement.
Results: Overall, this review discusses the diverse aspects of cannabis research developments, ranging from traditional use as herbal medicine to the latest potential in COVID-19, legal practices with updated patent status, and current state of the art genetic and genomic tools reshaping cannabis biotechnology in the modern agriculture and pharmaceutical industries.
Conclusions: Remarkable growth in genomic data, combined with fast-paced development of artificial intelligence (AI)-based data analysis tools have made it possible to explore the Cannabis plant at the genetic and molecular levels. In the future, the combination of these genetic technologies will make it possible to obtain enhanced expression rates, which will lead to enhanced cannabinoid yields in an economically feasible manner. Pharmacological research, coupled with rapidly evolving genome-based biotechnology, will further facilitate exploring the Cannabis plant for its tremendous potential in drug discovery.
Keywords: cannabis, cannabis research, plant biology, plant genetics, genomics
Introduction
Cannabis sativa L. is one of the earliest known cultivated plants since agricultural farming started around 10,000 years ago. (Schultes et al., 1974) It is a multi-purpose crop plant with diverse agricultural and industrial applications, ranging from the production of paper, wood, and fiber, to its actual and potential use in the medicinal and pharmaceutical industries. The first-ever report to reveal the prospects of C. sativa L. as a medicinal plant was published in 1843 and described the use of plant extracts to treat patients suffering from tetanus, hydrophobia, and cholera. (O'Shaughnessy, 1843) However, the first chemical constituent identified was oxy-cannabis, in 1869 (Bolas and Francis, 1869). Cannabinoids were being isolated as early as 1896, followed by a variety of full identifications like:
- cannabidiol (CBD) in 1940 (Jacob and Todd, 1940),
- tetrahydrocannabinol (THC) in 1964 (Gaoni and Mechoulam, 1964; Santavý, 1964),
- cannabigerol (CBG) in 1964 (Gaoni and Mechoulam, 1966), and
- cannabichromene (CBC) in 1966. (Gaoni and Mechoulam, 1966)
Identification of THC later led to an understanding of the endocannabinoid system, followed by the discovery of the first cannabinoid receptor (CB1) in 1988. (Devane et al., 1988; Russo, 2016). The CB1 receptor acts as a homeostatic regulator of neurotransmitters for pain relief mechanisms, but the same mode of action was responsible for the intoxicating effects from excessive cannabinoids use. This greater understanding of the mode of action of the CB1 receptor raised concerns about the adverse effects of cannabis use. Consequently, the plant was removed from the "medicinal" category and re-categorized exclusively to the category of "illicit drug."
Cultivation and use of the Cannabis plant for recreational, medical, and industrial use were strictly banned, which severely limited the scientific research in the field. Owing to strict legal regulations, the plant remained unexplored for its incredible potential in drug discovery for an extended period until it was legalized for medical use first in California and later in many countries around the globe. Extensive research followed legalization in order to explore the chemodiversity of cannabinoids for potential clinical value. In total, more than one thousand compounds have been identified, including 278 cannabinoids, 174 terpenes, 221 terpenoids, 19 flavonoids, 63 flavonoid glycosides, 46 polyphenols, and 92 steroids—have been identified. (ElSohly and Slade, 2005; Gould, 2015; Radwan et al., 2017) Nearly 278 of these compounds are cannabinoids and classified as phytocannabinoids (plant-based) to distinguish them from endocannabinoids (non-plant). Cannabimimetic drugs binding to CB1 receptors in the endocannabinoid system can also be found in algae, bryophytes, and monilophytes. (Carvalho, 2017; Kumar et al., 2019) The major cannabinoids in cannabis include THC, CBD, and CBC, as well as their precursors CBG and cannabinol (CBN). (Flores-Sanchez and Verpoorte, 2008) To date, 10 CBN-type, 17 CBG-type, 8 CBD-type, and 18 THC-type cannabinoids have been isolated. (Gaoni and Mechoulam, 1964) Cannabigerolic acid (CBGA), a CBG-type cannabinoid, is the central precursor for the biosynthesis of psychoactive THC, non-psychoactive CBD, and CBC. (ElSohly and Slade, 2005; Gould, 2015; Radwan et al., 2017)
Cannabinoid biosynthesis in plants occurs in specialized biosynthetic organs called glandular trichomes (Happyana et al., 2013) on female flowers and leaves. Several studies use metabolic profiling of trichomes to demonstrate variation in trichome size, density, and relative concentration of cannabinoids. (Happyana et al., 2013; Small and Naraine, 2016) However, the genetic mechanisms underlying the developmental changes in trichomes and consecutive cannabinoid content are still unknown. Apart from natural and chemical biosynthesis methods (Bovens et al., 2009), heterologous biosynthesis of cannabinoids has also been reported. (Luo et al., 2019) However, the considerable amount of side products is still one of the major bottlenecks in cannabinoid production. (Luo et al., 2019; Thomas et al., 2020)
This review highlights the latest research developments and challenges in Cannabis plant sciences, as well as the role of trichomes as biosynthetic sites, with a special focus on plant biology. Additionally, we discuss the existing legal practices with patent information for C. sativa L. We also discuss the new potential use of cannabinoids for COVID-19 treatment. Finally, we address the available genomic and transcriptomic resources and discuss their potential toward the genetic improvement of cannabis. Overall, we provide the first in-depth review of diverse aspects of C. sativa L. from traditional medicinal use to genomics insights and research perspective to broad industrial applications.
Methods
Cannabis-related publications were searched in four major scientific literature and citation databases of biomedical and life sciences journals: EMBL-EBI's Europe PMC (Data S1, Supplemental information), Elsevier's Scopus (Data S2, Supplemental information), National Library of Medicine's PubMed Central (Data S3, Supplemental information), and Clarivate's Web of Science. The search criteria—“cannabis OR marijuana OR hemp OR cannabinoids OR cannabidiol OR cannabinol”—were used to examine available research articles. Some 80,979 (EuropePMC), 64,637 (Scopus), 43,182 (Web of Science), and 28,759 (PubMed Central) cannabis-related research articles were found.
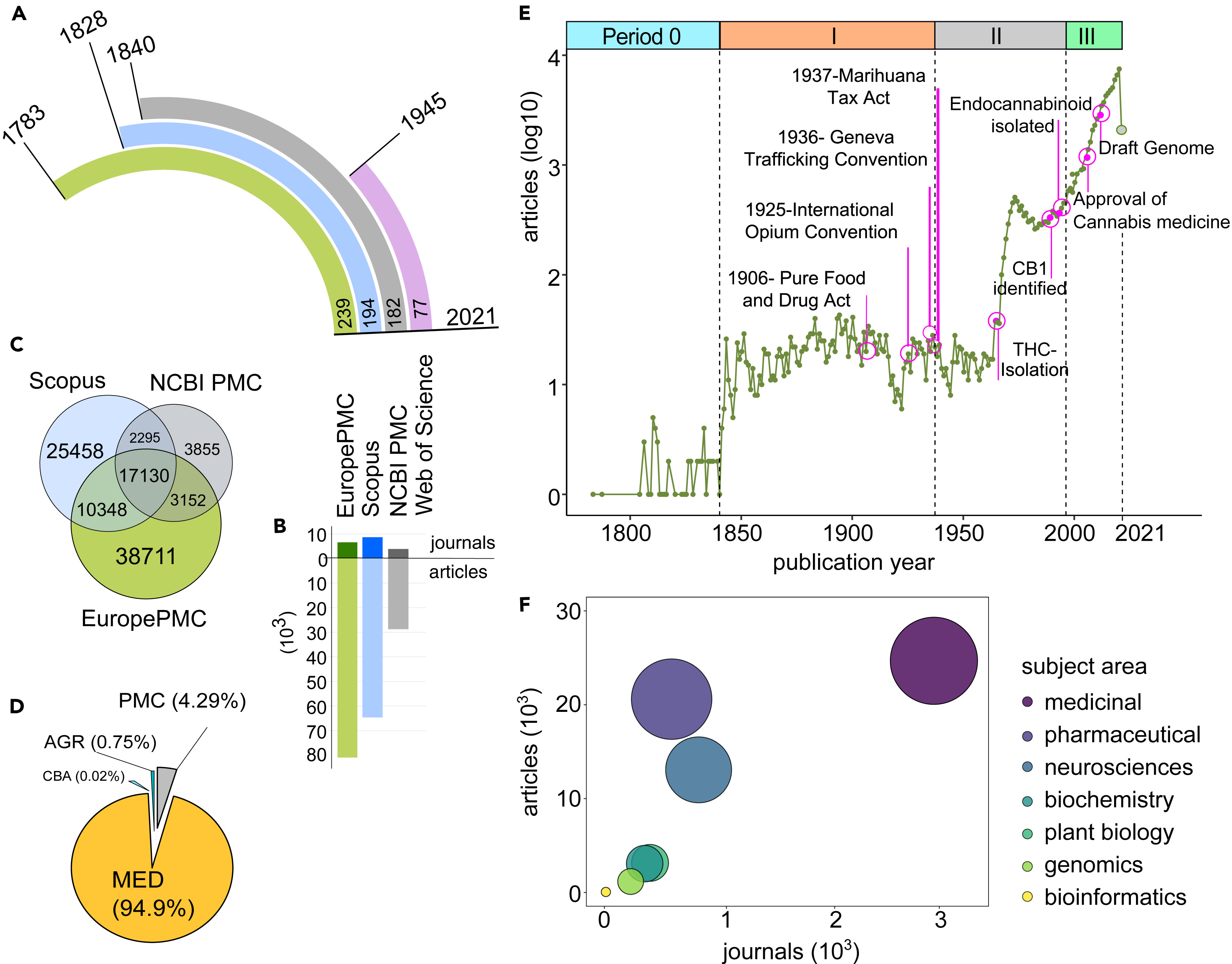
The sheer difference in the number of articles could be attributed to the years for which the cannabis records are present in the databases. Europe PMC currently holds cannabis records dating back 239 years, with the oldest publication dating to 1783, whereas Scopus has data for 194 years (dating back to 1828), Pubmed Central 182 years (dating back to 1840), and Web of Science 77 years (since 1945) (Figure 1A). Despite cannabis records only going back 77 years, the Web of Science record count exceeds Pubmed Central's, owing to a data acquisition policy similar to Scopus, wherein all the cited references for a publication are pulled and listed in the database.
Another major reason for the different records in the archives could be owing to the source repositories and partner journals. Although Pubmed Central has only 6.9 million articles from over 10,656 journals (as of April 2021), Scopus has more than 77.8 million records from nearly 23,500 journals, and Web of Science comprises over 171 million records, including journals, books, and proceedings. However, Europe PMC acquires data from multiple bibliographic repositories such as PubMed, MEDLINE, Pubmed Central, AGRICOLA, and Chinese Biological Abstracts (CBA) (Figure 1D). It includes more than 45.6 million documents, including articles, books, preprints, patents, conference papers, and microPublications.
Cannabis citation metadata was publicly available for bulk download from Europe PMC (6,586 journals), Scopus (8647 journals), and Pubmed Central (3864 journals) (Figure 1B). Among the various article identifiers used such as DOI, PMCID, and PMID, the DOI was found for 85.62% of records at Europe PMC, 85.44% of records at Scopus, and 91.9% of records at Pubmed Central. Since DOI was the only common identifier, it was used for the comparison of three datasets (Figure 1C). Cannabis records in Europe PMC comprised nearly 76.73% of Pubmed Central and 49.75% of Scopus data (Figure 1C). Hence, metadata from Europe PMC was selected for downstream bibliometric analysis. The majority of cannabis-related records in Europe PMC were from MEDLINE (94.94%), followed by 4.29% from Pubmed Central, with only 0.75% from Agricola and 0.02% from CBA (Figure 1D). The distribution of source databases indicates the most explored field in cannabis research for the last 239 years.
|
Trends of cannabis research from 1783 to 2021
C. sativa L. originated in central Asia and later spread to Europe during its cultivation, with diverse applications. Archaeological evidence of early medical use was found in fossil records dating back to 315–392 A.D. (Zias et al., 1993) Researchers largely have a consensus that the plant has at various times been used as traditional medicine. (Bridgeman and Abazia, 2017)
Based on our search results, spanning more than two centuries, we divide the scientific era into four periods (Figure 1E). Period Zero (1783–1840) marked the first-ever mention of Cannabis as a category of medicinal plant, in the years 1783 (Laurentius Crellius and Huntero, 1783) and 1787. (Wright et al., 1787) According to Europe PMC results, there were only 52 articles and 38 reviews in the next five decades. Most reports mentioned the botanical aspects of hemp and the quality of its fiber, with few observations about its use in traditional medicines.
Period I (1840–1937) began with the detailed evidence-based report of the chemical properties and medicinal potential of Cannabis indica (hemp) by William O'Shaughnessy (O'Shaughnessy, 1843), followed by an array of medicinal reports in articles from 1923. We identified 183 reviews in the subsequent 96 years. Scientific endeavors to experiment, observe, and understand the diverse medicinal applications of cannabis were still in the early stages. However, the 1900s witnessed a series of legal regulations in the direction of the criminalization of cannabis. Cannabis was starting to be categorized into lists of narcotic drugs, and "poisons rules," including the Pure Food and Drug Act of 1906, pushed for stricter measures for cannabis distribution. Later, the Second International Opium Convention of 1925 called for measures to regulate Indian hemp. Exports, unless exclusively for medical or scientific purposes or European hemp (for fiber), were banned. The Uniform State Narcotic Drug Act of 1925 and the Geneva Trafficking Conventions of 1936 resulted in criminalizing the cultivation, possession, manufacture, and distribution of cannabis derivatives. The Marihuana Tax Act of 1937 levied heavy taxes on the possession and selling of cannabis, excluding medical, and industrial use. As a consequence, the cultivation and procurement of cannabis for research purposes became increasingly difficult, severely limiting the research of medicinal cannabis during this era (see Figure 1E: Period I).
During Period II (1937–1996), cannabis research suffered major restrictions, owing to legal regulations in the first two decades, until the identification of the first cannabinoid—cannabidiolic acid or CBDA—in 1954 (Hanuš et al., 1975; Krejčí and Šantavý, 1955; Krejčí et al., 1958; Santavý, 1964) and the isolation of the most psychoactive component of cannabis—THC—in 1964. (Mechoulam et al., 1964) Other discoveries paved the way for decriminalization laws, including isolation of THC, discovery of the CB1 (Devane et al., 1988) and CB2 (Munro et al., 1993) receptors, and the emergence of the Compassionate Investigational New Drug program in 1978. The discovery of endocannabinoids and a growing understanding of the potential role of cannabis in the medicinal field also played a role during this period. (Hanus, 2009; Kabelik and Santavy, 1955) A significant uptick in published cannabis research was observed during this period, with 445 cannabis-related articles and 25 reviews being found between 1937 and 1964, ramping up to 8,888 articles and 773 reviews between 1964 and 1996 (see Figure 1E: Period II), although with a notably short period of decline in publications between 1973 and 1982.
Finally, Period III (1996-2021) began with the historical Compassionate Use Act of 1996 in California approving medical cannabis. Post-legalization (1996 onwards), cannabis has been extensively explored for its diverse potential in the pharmaceutical and medicinal industries. During Period III, cannabis research witnessed unprecedented growth, with nearly 67,777 articles, 13,202 reviews, and 493 preprints showing up in Europe PMC, of which 97.01% articles were published since 2000 (see Figure 1E: Period III). Approval of the first cannabis-based inhaler spray in 2005 (Perras, 2005; Pain, 2015) and publication of the first draft of the cannabis genome in 2011 (van Bakel et al., 2011) in this era were the two major accomplishments that exponentially accelerated research development.
The trends of cannabis study in the diverse array of research articles and journals indicate the core interests of the scientific community. To further investigate the most researched field, the journals of cannabis articles were categorized into scientific and social areas. The journals related to social-, legal-, and policy-based studies were merged into the subject category of "social research." The majority of broad science-based subjects were grouped into the following seven major categories: (i) medicinal, including all medical and medicinal subjects); (ii) pharmaceutical, comprised of pharmacology, pharmaceuticals, drug, toxicology, and chemical studies; (iii) neurosciences, comprised of neurological, brain-related, psychiatry, psychology, and cognitive studies; (iv) biochemistry, including biotechnology, microbiology, immunology, virology, and biochemistry; (v) genomics, including grouped genetic and genomic studies; (vi) plant biology, including plant sciences, agricultural, botanical aspects, plant-based pathogens, and environment studies; and lastly, (vii) bioinformatics, including data analytics. Journals that could not be classified into social research or science-based categories were excluded from downstream evaluation.
The science-based subject areas (74.47% of journals) were further compared for the corresponding number of articles and journals (Figure 1F). A distinct pattern was observed for the clinical aspects of cannabis, which remained a major focus from the very beginning. Some 94.76% of published articles addressed some sort of clinical aspect, including 64.51% articles addressing medicinal topics, 19.55% addressing pharmaceutical science topics, and 10.70% addressing the neurosciences. In contrast, plant biology and agricultural sciences comprised only 2.62% of articles, followed by 0.71% on genomics, and 0.07% on bioinformatics-based cannabis research. Genomics and bioinformatics are relatively new subjects, growing at a faster pace since the release of the first Cannabis draft genome in 2011. Recent advances in sequencing technologies have further propelled genomic and transcriptomic studies, with the purpose of dissecting the regulatory networks. The growth of genomic data in the public space has been met with the fast-paced development of bioinformatics tools for data analysis. In addition, the ongoing development of genomic tools using machine learning (ML) and artificial intelligence (AI) will facilitate improved genetic-level understanding of cannabis metabolism for the selective breeding of genetically modified cannabis with improved metabolic traits.
Cannabis sativa L. physiology and legal status
Physiological, morphological, and developmental aspects of the Cannabis plant are key in understanding its growth patterns and chemical profiles. However, plant growth and function are substantially influenced by abiotic factors and nutrient availability. Botanical aspects (Farag and Kayser, 2017), plant architecture, and florogenesis of female C. sativa plants (Spitzer-Rimon et al., 2019) with detailed trichome morphogenesis (Hammond and Mahlberg, 1977) has provided crucial insight into plant biology. However, it has also become increasingly important to determine the effect of abiotic factors on Cannabis growth and chemical yield, especially for large-scale commercial breeding programs. Hence, in-depth analysis of the effect of soil fertilization, salinity, temperature, and light conditions, as well as nutrient and water-use efficiency is key in establishing industrial-scale systems for the cultivation of hemp and marijuana varieties.
The first available records about the mineral nutrition of hemp plants were published by Tibeau et al. in 1936. (Tibeau, 1936) Later, in 1944, Clarence H. Nelson published the effect of varying soil temperature on hemp growth. (Nelson, 1944) The first publication with a detailed response of greenhouse cultivated cannabis to nitrogen (N), phosphorus (P), and potassium (K) was published in 1977. (Coffman and Gentner, 1977) Furthermore, two parallel reports by van der Werf et al. in 1995 discussed the impact of nitrogen fertilization on sex expression in hemp (van der Werf and van den Berg, 1995), and the effect of temperature on leaf and canopy formation. (van der Werf et al., 1995) Importantly, most physiological studies in Period II and Period III (Figure 1A) were published for hemp, with a focus on photosynthetic response and biomass yield under varying conditions such as temperature, water availability, nitrogen, and mineral nutrition. (Amaducci et al., 2002; Aubin et al., 2015; Finnan and Burke, 2013; Papastylianou et al., 2018; Tang et al., 2017, 2018) However, the first study to assess the chemical response of hemp plants wasn't published until 1997. (Bócsa et al., 1997)
The physiological response of drug-type medical Cannabis plants may differ from hemp plants, owing to the distinct genetic and chemical differences. Hence, a clear understanding of optimum factors for medical cannabis is inevitable for the efficient cultivation of plants with desired chemical composition. The first few studies that addressed medical cannabis and its photosynthetic response to photon flux densities, temperature, and CO2 conditions were published by Chandra et al. in 2008 and 2011. (Chandra et al., 2008, 2011) Bernstein and others further addressed the growth and chemical response of medical cannabis to mineral nutrition, especially N, P, and K. (Saloner and Bernstein, 2020; Shiponi and Bernstein, 2021; Saloner et al., 2019; Bernstein et al., 2019) Saloner and Bernstein (Saloner and Bernstein, 2020) reported optimum N concentration at 160 mg L−1, while N with lower levels showed several symptoms inducing necrosis and growth retardation and N with higher levels impacted in reducing concentrations of THCA and CBDA. Shiponi and Bernstein (Shiponi and Bernstein, 2021) showed a negative association of cannabinoid concentrations and yield with increasing P supply. Saloner et al. (Saloner et al., 2019) further determined the genotype-dependent effect of K nutrition on medical cannabis, reporting 240 ppm K detrimental for the genotype Royal Medic and stimulant for the Desert Queen genotype, while 15 ppm K was insufficient for both genotypes. Further, in 2019, Bernstein et al. (Bernstein et al., 2019) discussed the combined effect of NPK nutrition upon cannabinoid concentration.
In addition to soil nutrients, the heavy metals uptake potential of hemp varieties has also been thoroughly investigated by multiple reports in the 2000s. (Citterio et al., 2003; Ferrarini et al., 2021; van Ginneken et al., 2007; Hoseini et al., 2012; Sakizadeh et al., 2016; Shi and Cai, 2010; Rheay et al., 2021; Vandenhove and Van Hees, 2005) Industrial hemp varieties of C. sativa have also been shown to grow well in soils contaminated with heavy metals (Citterio et al., 2003; van Ginneken et al., 2007; Hoseini et al., 2012; Sakizadeh et al., 2016; Shi and Cai, 2010; Vandenhove and Van Hees, 2005) and reported for their heavy metal accumulation. Several field projects have assessed the phytoremediation potential of hemp plants for the reclamation of contaminated and radioactive soils. (Ferrarini et al., 2021; Rheay et al., 2021)
Cannabis cultivars are classified into drug-type (marijuana), fiber-type (hemp), and neutral (zero cannabinoid) plants with distinct cannabinoid constitutions. Drug-type cultivars with THC/CBD ratio ≥10 are classified as chemotype I, while those with THC/CBD ratio ranging from 0.2 to 10 are grouped as chemotype II. In contrast, fiber-type cultivars with THC/CBD ratio <0.2 are categorized as chemotype III. Chemotype IV also has low THC contents but with the potent percentage of CBG. Furthermore, the chemotypes producing very little to almost zero cannabinoid compounds (neutral) are grouped as chemotype V (Cascini et al., 2012; Hartsel et al., 2016) and were first described by Mandolino et al. in 2004. (Mandolino and Carboni, 2004). Apart from cannabinoid (THC, CBD) content, drug and fiber-type plants have significant genetic variation. Sawler et al. (2015) described that marijuana is genetically inclined toward sativa, and hemp has a similarity with the indica type. (Sawler et al., 2015) Moreover, each plant type has unique applications, differentiating them from each other. For example, the fiber-type "hemp” plant has mostly food and industrial applications, including production dietary products, hemp oil, seeds, and fiber, while the “marijuana” drug-type plant is used exclusively for medicinal and recreational purposes.
Despite such a huge genetic and application diversity, both types of cannabis plants were placed under Schedule I of the Controlled Substances Act in 1970. (Drug Enforcement Administration, 1970) These restrictions had a serious impact on cannabis-related research, preventing the scientific community from studying the potential of diverse yielding traits for hemp. However, after 44 years, Section 7606 of the Agricultural Act of 2014 finally distinguished hemp from marijuana.[1] Approval of the law, along with the 2018 Farm Bill[2], opened the window for the scientific community in the U.S. to conduct research and cultivate hemp. Since then, most U.S. states[3] and more than 47 countries around the world have been growing hemp for research and industrial use. (Schluttenhofer and Yuan, 2017) On the other hand, marijuana research and legalization have been expanding at a comparatively slower rate, and till now only 16 countries have legalized medicinal cannabis. (Aguilar et al., 2018) Furthermore, a detailed study would be desirable to understand the gene function, the genetic composition, and the underlying mechanisms regulating the diversity of cannabinoids in both major varieties. Availability of the regeneration protocol (Lata et al., 2016) and transformation studies (Schachtsiek et al., 2018) could be utilized for the expression studies to unravel the mystery of these mechanisms, especially in trichomes.
Trichomes and cannabinoid biosynthesis
Glandular trichomes are the primary site for cannabinoid biosynthesis and accumulation (Lanyon et al., 1981) in C. sativa. The biosynthesis of cannabinoids (Andre et al., 2016; Degenhardt et al., 2017) starts from the plastidial localized methylerythritol 4-phosphate (MEP) pathway, resulting in the formation of geranyl pyrophosphate (GPP) (Marks et al., 2009) and the fatty acid pathway leading to the production of olivetolic acid (OA). (Raharjo et al., 2004) GPP and OA in the presence of olivetolic acid cyclase (OLS) (Gagne et al., 2012; Stout et al., 2012) and an aromatic prenyltransferase catalyze the reaction to form CBGA (Gagne et al., 2012; Fellermeier and Zenk, 1998; Taura et al., 2009), which is the central precursor for cannabinoid biosynthesis. In 2011, van Bakel et al. analyzed the transcriptomic and genomic data and described the exclusive presence of the THCAS and CBDAS in the drug and hemp-type plant, respectively. (van Bakel et al., 2011) It is suggested that the activation of respective enzymes from the central precursor CBGA is responsible for regulating the THC and CBD concentration for each chemotype. However, the precise regulatory mechanism is still unknown.
Besides biosynthesis, understanding the trichome physiology is also vital to elucidate the trafficking and localization of metabolites. For cannabinoid biosynthesis, there exist three major reactions:
- biosynthesis of monoterpene precursor (GPP) via MEP and fatty acid intermediate (OA) from polyketide pathway,
- prenylation of the precursors, and
- cyclization.
The MEP pathway in plastid prenylation is localized in the chloroplast membrane, where the C-prenylated CBGA synthase is membrane-bound. The integration of the enzyme in the membrane seems essential, and the folding pattern is crucial for its functioning. Therefore, simple cloning and functional expression of this enzyme in a heterologous host such as yeast to generate the desired cannabinoids is challenging. Terpenoid cyclization reactions are the most complex reactions found in nature, and the biotransformation from CBGA to THCA by the THCA synthase is assumed to occur in the cytosol. This hypothetical model is under ongoing debate, and it might be likely that biocatalysis occurs in the extracellular oil container under a non-aqueous environment. (Lange et al., 2015) In 1992, Mahlberg and Kim postulated that THCA synthase is located in the outer membrane of the head cells or even attached on the outer membrane surface extending into the essential oil. (Mahlberg and Kim, 1992) In recent studies, Liquid chromatography–tandem mass spectrometry (LC-MS/MS) was used to detect a functional, active THCA and CBGA synthase in the exudates from glandular trichomes of cannabis. (Rodziewicz et al., 2019) Zirpel et al. described the need for an excellent understanding of protein chemistry and folding of these enzymes to produce the cannabinoid using a heterologous host. (Zirpel et al., 2018) Detailed knowledge of genetic regulatory mechanisms underlying cannabinoid biosynthesis is a future challenge. Identification of regulatory elements such as transcription factors (TFs) and microRNAs (miRNAs) could be utilized to understand the mechanistic insights of trichomes initiation, development, and densities. An in-depth understanding could be applied toward the breeding of genetically improved cannabis varieties with enhanced cannabinoids concentration in trichomes.
Developments in cannabis genomics
Drug- and fiber-type plants differ in biosynthesis, concentration, and composition of metabolites. (Finnan and Burke, 2013) To determine the genetic variations regulating plant-specific differences, it is essential to compare the genomes. Advanced sequencing technologies, combined with continuously improving bioinformatics tools, have allowed rapid sequencing and analysis of multiple genomes and transcriptomes. The very first draft genome of C. sativa was released in 2011 by Bakel et al. (van Bakel et al., 2011) They sequenced the marijuana cultivar Purple Kush by using Illumina short reads and assembled them in combination with 454 reads. They also sequenced the fiber-type hemp cultivar Finola for a genome-level comparison. In addition to whole genome sequencing, the first complete mitochondrial reference genome was also obtained in 2016 from the cannabis hemp variety Carmagnola. (White et al., 2016) Later, in July 2016, two complete chloroplast genomes of marijuana (THC-dominant) African variety Yoruba Nigerian and Korean hemp non-drug variety (low THC) Cheungsam (Oh et al., 2015) were sequenced and used to determine the phylogenetic position of C. sativa relative to other members in the order Rosales. Furthermore, Vergara et al. released the complete chloroplast genomes of two Cannabis hemp varieties, the Carmagnola (Italian) and Dagestani (Russian), in September 2016 to determine their genetic distance compared with the closest Cannabaceae chloroplast of Humulus lupulus variety Saazer. (Vergara et al., 2016)
Increasingly growing support for open data policies by multiple industries is improving transparency in cannabis agriculture. In 2016, the industrial lead in cannabis research from Courtagen Life Sciences and Phylos Bioscience independently generated the genomes of hybrid marijuana strain (THC-dominant) Chemdog91 (by Illumina GAII) and marijuana strain (CBD-dominant) Cannatonic (using PacBio), respectively. Phylos Bioscience also released genomic data on 850 cannabis strains as a part of the “Open Cannabis Project” for plant breeding programs. With an objective to explore cannabis population genetics, Phylos Bioscience developed a three-dimensional interactive map of nearly 1,000 cannabis strains.[4] In 2017, the genome of hybrid marijuana cultivar Pineapple Banana Bubba Kush (PacBio) was released as part of the Cannabis Genomic Research Initiative. In 2018, Grassa et al. generated the first chromosome-level assembly for the genome of CBDRx, a high-CBD cultivar of C. sativa by using advanced long-read Oxford Nanopore Technology (ONT) and PacBio Single-Molecule Real-Time (SMRT) sequencing. (Grassa et al., 2018) In 2019, Laverty et al. improved the initial draft assemblies (van Bakel et al., 2011) of drug-type Purple Kush and hemp-type Finola to the chromosome-level by using ultra-long PacBio reads. (Laverty et al., 2019) In addition to genomes of high-CBD and -THC marijuana and hemp cultivars, a medicinal cannabis strain with a balanced THC/CBD ratio was sequenced by Shivraj et al. (Braich et al., 2020)
Until 2020, nearly all cannabis genomes had been obtained from the hemp and marijuana cultivars, selectively bred for generations. However, cultivars lose genetic diversity owing to domestication and successive plant breeding for selected traits. In contrast, the wild-type genomes exhibit relatively high heterozygosity and genetic diversity, which might provide unique evolutionary insights into the cannabis genome. In 2020, Gao et al. sequenced the first samples of C. sativa wild-type “Jamaican Lion,” growing in the geographically isolated Himalayan region in Tibet. Because these wild-type plants retained the ancestral genetic make-up, the data generated from this study was used as a tool to determine the inheritance patterns and evolutionary inference of cannabis. (Gao, 2020)
The published genomes of high-THC and -CBD marijuana cultivars, as well as hemp varieties, have exhibited inconsistent chromosomal nomenclature, arrangement, and a varying degree of gaps. By the end of 2020, Shivraj Braich et al. had generated a relatively complete draft genome assembly for Cannbio-2, the medicinal cannabis strain with a balanced THC/CBD ratio. (Braich et al., 2020) To present date, only 13 cannabis genomes are publicly available at the National Center for Biotechnological Information (NCBI). Of those 13, three assemblies are at the chromosome level, seven at the contig level, and one at the scaffold level. However, as of March 2021, the 1000 Cannabis Genomes Project comprises genomic data of some sort for nearly 1,000 samples from multiple cannabis strains. These datasets were the first genome data released on the Google Cloud Big Query database.
The continuous expansion of the list of cannabis genomes needs collaborative efforts toward curating the information. As such, academic and industry experts in diverse fields formed the International Cannabis Research Consortium (ICRC) during the annual PAG meeting in 2020. Despite several cannabis genome assemblies, the selection of a single standard reference genome is still a huge challenge for the scientific community, especially plant breeders. However, the ICRC proposed the CBDRX Cs10 assembly as the most complete reference for use in cannabis genome research. (Grassa et al., 2021) Additionally, a member genomics company, NRGene, generated an integrated Cannabis and Hemp Genomic Database (CannaGENE) optimized and curated for the genomics-based breeding of cannabis varieties. Finally, in 2021, the first-ever open-access and comprehensive database of cannabis genome, the CannabisGDB, was released (Cai et al., 2021) with integrated bioinformatics tools for the analysis of datasets.
Overall, the genomic data of diverse cannabis genotypes represent untapped reservoirs of genetic information which could be applied toward pan-genomic understanding of cannabis evolution and determining the effect of genetic variations upon the pathways, development, and concentration of cannabis derivatives. A detailed genetic atlas would facilitate the designing and further breeding of cannabis varieties for preferred metabolic yields.
Developments in cannabis transcriptomics
The availability of several high-quality cannabis genomes made it easier to apply the transcriptome sequencing to elucidate detailed expression dynamics in a time-, tissue-, stage-, and chemotype-dependent manner. Furthermore, the differential expression analysis provides in-depth insight into co-related gene networks. In 2011, Bakel et al. sequenced and compared the transcriptomes of marijuana variety Purple Kush (PK) and hemp cultivars Finola (FN) and USO-31. Gene expression analysis revealed preferential expression of cannabinoid and precursor pathway-associated genes in marijuana (PK). Expression of THCA synthase in the PK and CBDA synthase in FN was found to be consistent with the exclusive production of psychoactive THC in marijuana. In a recent study, transcriptomics of hemp-type plants was analyzed to determine the expression profile of the fiber-type plant at the various developmental stages. (Guerriero et al., 2017) Similarly, the transcriptome of marijuana flowers at different stages was captured and sequenced, notably with a gene expression pattern consistent with its cannabinoid contents. (van Bakel et al., 2011)
As glandular trichomes are the central reservoir for cannabinoids (Lanyon et al., 1981; Turner et al., 1981), the trichome transcriptome could yield valuable insight to determine the variation in cannabinoid biosynthesis, composition, and concentration between the drug- and fiber-type plants. Importantly, the identification of the differentially expressed genes could unravel the underlying molecular mechanisms of natural genetic and metabolic variation. The gene expression in trichomes of female plant strain Cannobio-2 was compared with genome-wide transcriptomics of female floral tissues at different stages of development, as well as other tissues including female and male flowers, leaves, and roots. (Braich et al., 2019) The extensive-expression atlas was applied toward the identification of genes expressed preferentially in various tissues at different developmental stages. Interestingly, the majority of genes involved in terpenoid and cannabinoids synthesis were significantly overexpressed in trichomes. In 2021, Grassa et al. used genomic and expression-associated expression of THCAS and CBDAS with THC:CBD ratio by Quantitative trait Loci (QTL) analysis of cannabis cultivars. (Grassa et al., 2021)
Datasets from similar genomics, transcriptomics, microbiome, and metagenomics studies of various cannabis strains are currently accessible from the Sequence Read Archive (SRA) repository at NCBI. In the past three years, there has been unprecedented growth in cannabis genome and transcriptome studies, as well as corresponding SRA entries. To date, there are over 4,571 BioSamples from multiple studies related to cannabis, of which 2,871 public BioSamples are exclusively for C. sativa, with 2,546 DNA and 325 RNA-Seq datasets in SRA. The SRA data for transcriptomics and metagenomics have been reportedly procured from various tissues, including seeds (3), flowers (116), leaves (138), shoot (13) stem (175), root (76), and trichomes (62), while genomic data lacks tissue-specific information. In-depth transcriptomic studies will be required in the future to improve the understanding of regulatory genetic networks.
Patents for Cannabis sativa L.
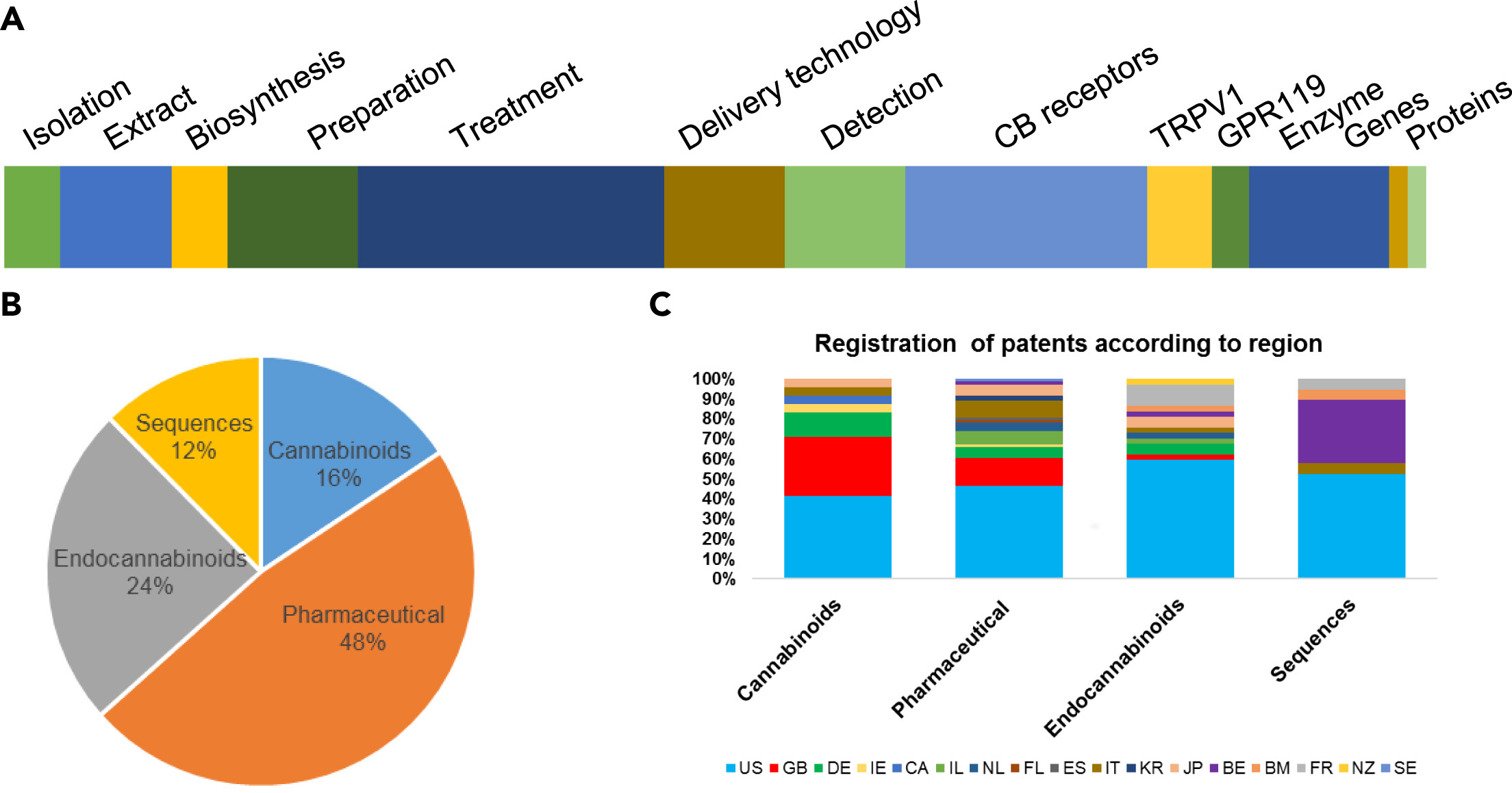
One of the fundamental aspects of patents, especially in medical science or biotechnology, is to involve industrial partners in investing in research and development. (Cook-Deegan and Niehaus, 2014) cannabis-related patents have been issued by the United States Patent and Trademark Office (USPTO) since 1942. More than 1,500 applications have been filed just in the USPTO. Among them, approximately 500 applications got patent protection rights (Weed, 2017), and most of them were from the last decade. The exponential increase in the number of patents shows the future potential for the growing cannabis industry. Here, we analyzed the patents spatiotemporally and categorized them into four main classes: (i) patents related to cannabinoids as constituents, (ii) pharmaceutical applications, (iii) endocannabinoid pharmacology, and (iv) genome- and gene-related. Among the suggested four categories, the patents related to the pharmaceutical application were the most significant category, with 73 patents registered. These are further sub-grouped into the (i) preparation of the drugs, (ii) treatment, (iii) delivery technology, and (iv) detection method, each with 14, 33, 13, and 13 patents, respectively. Others have examined cannabis-related patents as well. For example, Gerra et al. (2010) reviewed endocannabinoid-related patents, comprised of the CB1/2 receptor (26), TRPV1 (6), and GPR119 (4) in 2010. The category of cannabinoids consists of (i) cannabinoid isolation, (ii) extraction, and (iii) synthesis- or biosynthesis-related patents, each with 6, 6, and 12 patents granted, respectively. For the division of the sequences, 15 patents are from enzyme inhibition, followed by the gene and the protein, each with two patents. Most of the patents are from the US (49.6%), followed by Great Britain (11%) and a variety of European countries (Flores-Sanchez and Ramos-Valdivia, 2017) (Figure 2). In addition, 25 patents for fiber/textile, 10 for foodstuff, five for the paper industry, three for architecture, one for biofuel, and three for plant breeding have been registered. Also, four patents each in the category of oil, extracts, and cosmetics have been filed.
|
However, we have to keep in mind that a certain cannabinoid invention can be referred into more than one patent category. For instance, cannabinoids are highly hydrophobic by nature and thus they have low bioavailability in the human body. As a result, a new class of cannabinoid-glycosides has been created, whose representatives are produced through enzymatic glycosylation. This novel strategy led to increased aqueous solubility of the target cannabinoids and resulted in four patents (WO2017053574, US20190153461, US20190078168, and WO2020239784). Recently, a new method of producing one or more cannabosides by feeding an insect a cannabinoid was patented (WO2021146687). These new classes of cannabinoid glycosides generated vast structural diversity and have greatly improved water solubility, enabling new pharmaceutical formulations and multiple administration routes (oral, parenteral, or transdermal). The discovery of the genes encoding glycosyltransferases may belong to different categories of the cannabinoid patent family, that is, genes, enzymes, delivery technology, etc.
The exponential enhancement of the patent number during recent years in the diverse areas of cannabinoid applications is indicative of the increased commercial interest in this class of natural compounds. The various pharmaceutical applications will continue to shape primarily the path of future invention of cannabinoids.
Cannabis in COVID-19
C. sativa has become increasingly known for its potential anti-inflammatory properties. (Prakash et al., 2009) The severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) virus and its resulting disease, COVID-19, can result in life-threatening lung inflammation. A reduction of this inflammation is optimal, and means of reducing lung inflammation have been tested in the mice animal model. Interestingly, cannabinoid isolates such as CBD and THC have also been tested in humans, even long before the onset of the global pandemic owing to the spread of SARS-CoV-2. (Petrosino et al., 2018; Pellati et al., 2018; Almogi-Hazan and Or, 2020) Immune responses during severe cases of COVID-19 trigger the inflammation of human lung tissue, resulting in acute respiratory distress and failure. This immune response to the overproduction of pro-inflammatory cytokines is known as a cytokine storm. (Esposito et al., 2020) Respiratory distress from COVID-19-induced lung inflammation is the leading cause of the disease's high mortality rate. Phyto-cannabinoids, especially CBD, have exhibited a remarkable anti-inflammatory effect through CB2 inhibitory activity and an agonistic effect on the peroxisome proliferator-activated receptor γ (PPARγ). (Malinowska et al., 2021) Additionally, CBD, CBN, and THC have also been shown to exhibit some anti-viral effect against COVID-19 in cell-based assays, with the same potency as the standard clinical references (remdesivir and lopinavir). (Raj et al., 2021) However, the complete antiviral mechanism of cannabinoids against SAR-CoV-2 infection is still unknown. Therefore, detailed pharmacological research studies are urgently needed to explore the immunotherapy potential of cannabis against SARS-CoV-2 infection.
Conclusion and future prospective
Cannabis legalization fueled scientific research in cannabinoid compounds for their potential in medicinal, pharmaceutical, and neurological applications. However, with recent developments in sequencing technologies, there has been a paradigm shift in cannabis research toward the genomics of fiber- and drug-type plants. The remarkable growth in genomic data, combined with fast-paced development of AI-based bioinformatics and data analysis tools, has made it possible to explore the Cannabis plant at the genetic and molecular levels. Integrated omics studies combining genomic and expression data with metabolite profiles are now beginning to understand the genetic regulation of the cannabinoid biosynthesis pathway, especially by unraveling the association between the expression of cannabinoid genes with THC:CBD ratio and cannabinoid content. The knowledge could be further applied to genetically modified cannabis, with optimized pathways for preferred metabolite yield and composition. Advanced biotechnology methods could be further extended for recombinant production of cannabinoids in metabolically engineered hosts such as yeasts or bacteria. Currently, the recombinant production of THC in yeast is challenging, owing to unstable THCA and CBGA expression and high amounts of side products. However, in the future, the combination of genetic technologies to obtain enhanced expression rates will lead to enhanced cannabinoid yields in an economically feasible manner. In addition, cannabinoids have been recently shown to exhibit anti-inflammatory and immunosuppressing effects against the COVID-19 immune response. However, further evidence-based clinical studies are needed to determine the efficacy and safe dosage of cannabis extracts for treatment or prevention of COVID-19. Pharmacological research, coupled with rapidly evolving genome-based biotechnology, will further facilitate exploring the Cannabis plant for its tremendous potential in drug discovery.
Supplemental information
Acknowledgements
Author contributions
T.H. and O.K. designed the concept, while T.H. and G.J. performed a literature search and did bibliometric analysis. T.H. and G.J. wrote the sections “History of cannabis research, Trends in past two centuries of cannabis research and Development in Cannabis Genomic and transcriptomes”. T.H. an N.V. wrote the “patent” section. T.P. contributed in “Cannabis in covid-19” section. O.K. supervised the study and all the authors contributed to the final manuscript.
Conflict of interest
The authors declare no competing interests.
References
- ↑ Office of the Secretary; Drug Enforcement Administration; Food and Drug Administration (12 August 2016). "Statement of Principles on Industrial Hemp". Federal Register. pp. 53395-53396. https://www.federalregister.gov/documents/2016/08/12/2016-19146/statement-of-principles-on-industrial-hemp.
- ↑ Abernethy, A. (24 July 2019). "Hemp Production and the 2018 Farm Bill". U.S. Food and Drug Administration. https://www.fda.gov/news-events/congressional-testimony/hemp-production-and-2018-farm-bill-07252019.
- ↑ National Conference of State Legislatures (16 April 2020). "State Industrial Hemp Statutes". https://www.ncsl.org/research/agriculture-and-rural-development/state-industrial-hemp-statutes.aspx.
- ↑ "Phylos Galaxy". Phylos Bioscience, Inc. 2021. https://phylos.bio/galaxy/.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. References aren't listed in the order they appear in the original, but they do for this version, by design. In some cases important information was missing from the references, and that information was added. A mention of the U.S. 2018 Farm Bill and several citations related to that bill, as well as one in regards to Section 7606 of the Agricultural Act of 2014, were added. This version is otherwise unchanged in compliance with the "NoDerivatives" portion of the original's license.